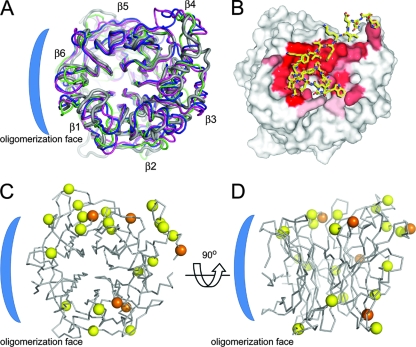
FIG. 5.
Conservation of N-linked glycosylation sites on NiV-G across paramyxoviruses. (A) Cartoon representation (Cα trace) of the crystal structures of the globular β-propeller domains from envelope attachment glycoproteins of NiV-G (gray; PDB accession number 2VSM), NDV-HN (pink; 1E8T), PIV3-HN (green; 1V2I), and SV5-HN (blue; 1Z4Y). (B) Residues from the G-H loop of EFNB2 (sticks; residues 115 to 127) in complex with NiV-G (van der Waals surface), where sticks are colored yellow for carbon, blue for nitrogen, and red for oxygen. The surface is colored by a gradient of red (completely buried) to white (not buried) according to the fraction of buried surface area. (C) Superimposition of Cα trace ribbon of NiV-G with N-linked glycosylation sites of NiV-G, HeV-G, PIV1, PIV2, PIV3, Sendai virus, and NDV mapped as spheres at the Cα on equivalent residues. The position of the NiV-G glycans (at Asn306, Asn378, Asn417, Asn481, and Asn529) are indicated by orange spheres, and the positions of glycans from the other viruses are indicated by yellow spheres. (D) View of NiV-G formed by a 90° rotation of panel C. The putative oligomerization face is indicated by a blue bracket.