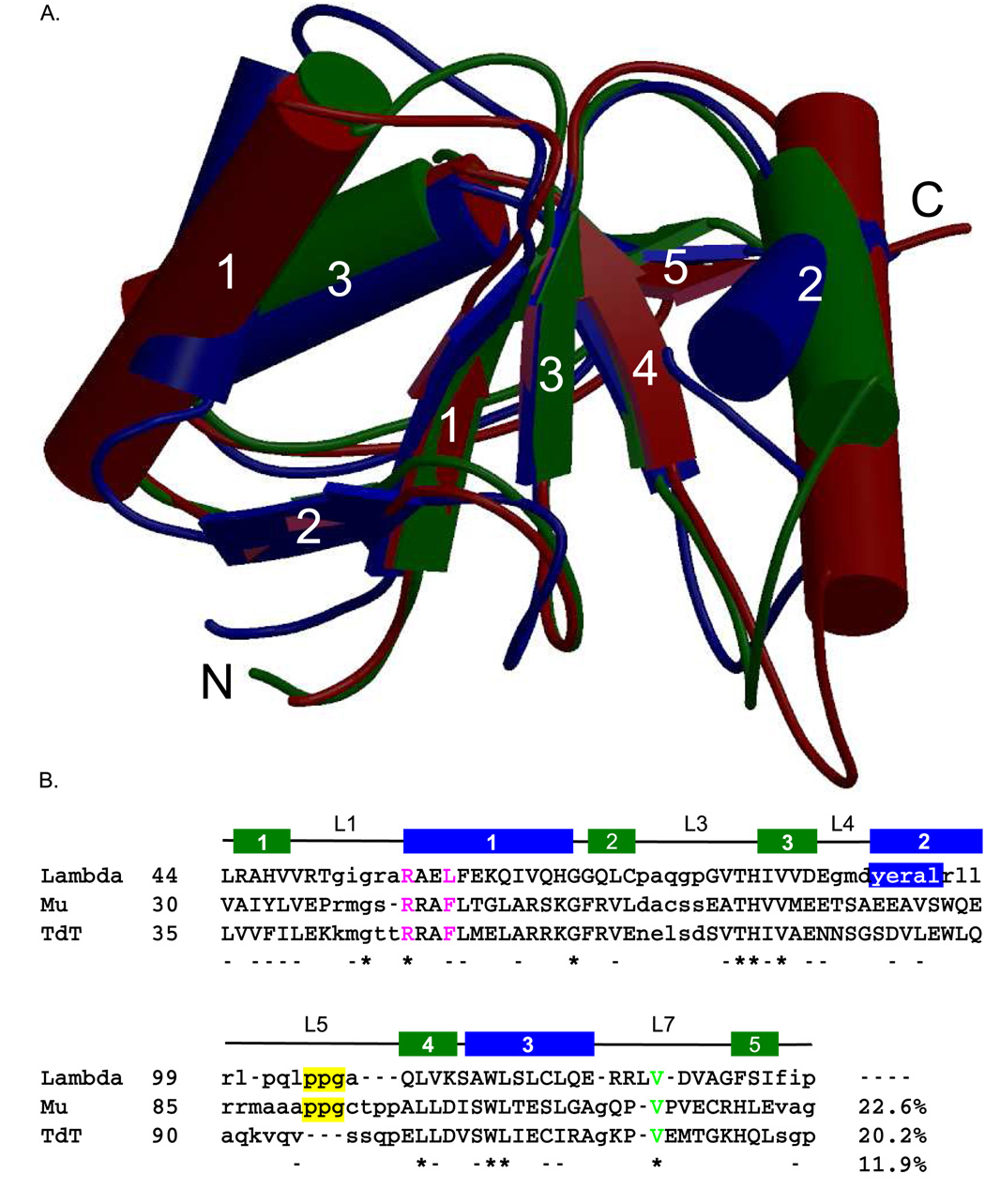
Figure 3.
Structural alignment of the BRCT domains of the Family X polymerases. (A). Superposition of the BRCT domains from human Pol µ (PDB code 2DUN, maroon), TdT (PDB code 2COE, green), and Pol λ (PDB code 2JW5, blue). α-helices (cylinders) and β-strands (directional arrows) are labeled numerically. This figure was created using MolScript [50] and Raster3D [51] (B). Structure-based primary sequence alignment. The positions of the conserved secondary structural elements of the Pol λ BRCT domain are shown as follows: β-strands are drawn as green boxes and α-helices are drawn as blue boxes. Structurally homologous residues are in capital letters, while nonhomologous regions are shown in lower-case letters. Absolutely conserved residues are indicated by asterisks below the alignment, and nearly conserved residues are dashed. The PPG motif is boxed in yellow. The conserved valine residue required for protein stability is shown in green. Residues mutated for biochemical analysis of binding to NHEJ protein partners are in magenta.