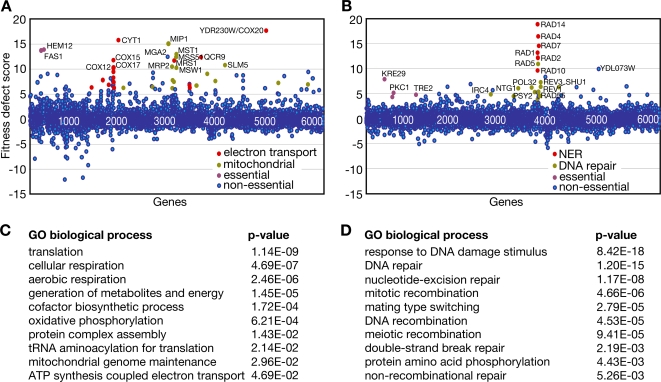
Figure 2. Chemical-genetic profiling of compound 13 and compound 15.
A. Identification of gene deletion mutants that confer sensitivity to 9 µM compound 13 by chemical-genetic profiling with the yeast heterozygous essential gene deletion mutants and the homozygous diploid non-essential gene deletion mutants. Fitness defect scores are calculated based on barcode microarray hybridization, and are plotted on the y-axis. Positive values indicate under-representation in the treated pool, and therefore sensitivity of the corresponding mutant. The heterozygous (first 1200 genes) and homozygous deletion mutants are arranged on the x-axis alphabetically. Essential genes (purple), electron transport genes (red) and genes annotated for mitochondrial function (yellow) in the top 50 hits are indicated. B. Identification of gene deletion mutants that confer sensitivity to 9 µM compound 15. Essential genes (purple), nucleotide excision repair genes (NER; red) and DNA repair genes (yellow) in the top 50 hits are indicated. C. Enrichment of GO biological processes in compound 13 sensitive strains with a fitness defect score greater than 4. D. Enrichment of GO biological processes in compound 15 sensitive strains with a fitness defect score greater than 4.