Figure 1.
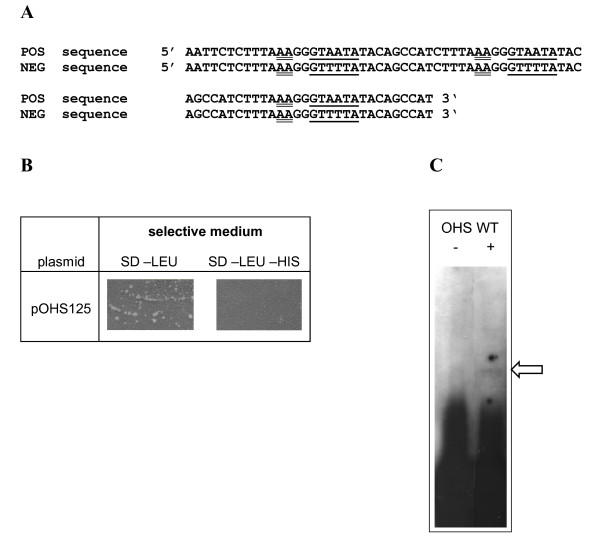
Identification of the CAE-binding protein by the One Hybrid System. (A) Sequences of oligonucleotides used for the in vivo yeast assay (POS; (GG)GTAATA; used to construct pHIS-CAE) and the negative control experiment (NEG (GG)GTTTTA; used to construct pHIS-mCAE). (B) Negative control experiment using pHIS-mCAE containing yeast strains transformed with pOHS125. Growth on synthetical dropout medium lacking leucine (SD-LEU; numerous colonies) indicates successful transformation, inability to grow on SD-LEU-HIS (empty plate, no colonies) confirms that due to the mutation of (GG)GTAATA to (GG)GTTTTA DNA binding of the protein encoded on pOHS125 is abolished and therefore sequence specific. A detail of the plates after the transformation is shown, equal amounts of transformation mix were plated on both positive and negative selection medium. (C) Electrophoretic mobility shift assay using 5 ng of labeled oneH-CAE as oligonucleotide and 5 μg of a GST-fusion protein with LIMPET aa457 – aa636 (+) which was functional in yeast. GST-elution buffer (see Materials and Methods) containing 20% glycerol and lacking protein was used as negative control (-; free probe). The complex formed is indicated by an arrow.