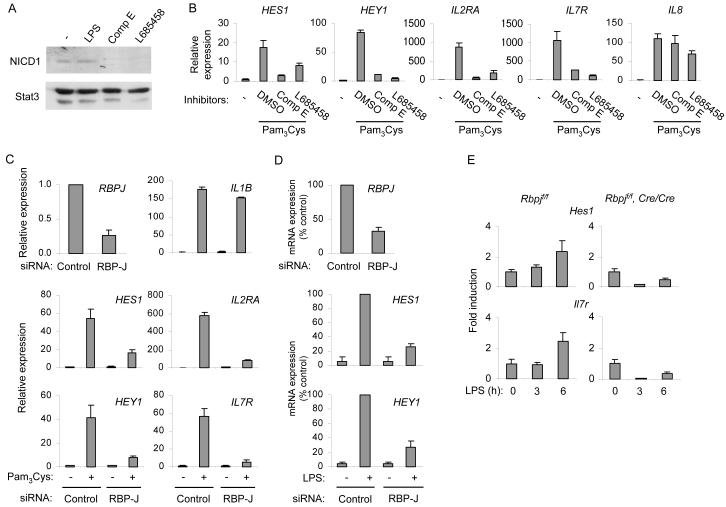
Figure 2.
Notch signaling is necessary for TLR induction of Notch target genes
(A) Human macrophages were activated with 10 ng/ml of LPS for 1 h (lane 2), or incubated with γ-secretase inhibitors Compound E (Comp E; 10 μM) or L685458 (5 μM) (lanes 3 and 4). Whole cell extracts were assayed for NICD1 by immunoblotting. Stat3 served as a loading control. One representative experiment out of two performed is shown.
(B) Human macrophages were pretreated with DMSO vehicle control or γ-secretase inhibitors for 2 d and subsequently stimulated with 10 ng/ml of Pam3Cys for 3 h. mRNA was measured by real time PCR. Data are shown as means + SD of triplicate determinants and are representative of five independent experiments.
(C, D) Primary human macrophages were transfected with control non-targeting or RBPJ-specific short interfering RNAs. 4 d post transfection, cells were stimulated with TLR ligands (10 ng/ml) for 3 h, and mRNA was measured using real time PCR. (C) Macrophages were activated with Pam3Cys. One representative experiment out of three performed is shown. Results are presented as mean + SD of triplicate determinants. (D) Macrophages were stimulated with LPS. Percentage of maximal expression was calculated relative to mRNA amounts in LPS-stimulated control cells and data are expressed as means + SD of three independent experiments.
(E) Bone marrow derived macrophages from Rbpjflox/flox (f/f) mouse and Rbpjf/f, Cre/Cre littermate controls were stimulated with 1 ng/ml of LPS for the indicated periods. RNA was extracted and mRNA was measured using real time PCR. Unstimulated controls within each genotype were set as 1. Data are shown as means + SD of triplicate determinants and are representative of three independent experiments. The difference in Hes1 expression between control and RBP-J-deficient macrophages at 6 h was statistically significant (P = 0.004 by paired Student’s t test).