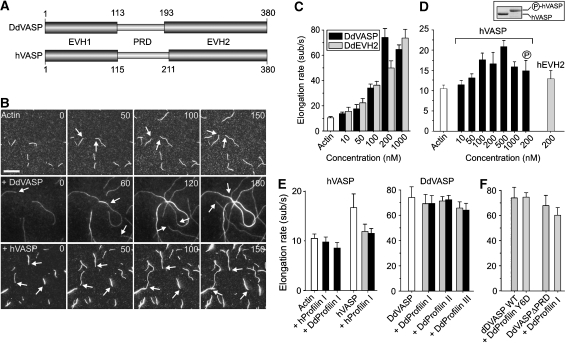
Figure 1.
Analysis of VASP-mediated actin assembly by TIRF microscopy. (A) D. discoideum DdVASP and human hVASP share a similar domain organization encompassing an N-terminal EVH1, a central proline-rich region (PRD) and C-terminal EVH2 domain. Numbers indicate amino-acid residues. (B) VASP accelerates actin assembly in vitro. A concentration of 1 μM ATP-actin and 0.3 μM Alexa-Fluor-488 labelled ATP-actin (from now on referred to as 488 actin) was polymerized in the presence or absence of either 200 nM DdVASP or 200 nM hVASP on NEM-myosin II-coated glass slides in 1 × TIRF buffer containing 10 mM imidazole, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 50 mM DTT, 15 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase and 0.5% methylcellulose (4000 cP), pH 7.4. Time-lapse micrographs of the assembly reaction are shown. The time is indicated in seconds on top. Arrows mark barbed ends during filament elongation. Scale bar, 10 μm. (C) Concentration dependence of the elongation rates of DdVASP-mediated actin assembly by the full-length protein and the EVH2 domain (DdEVH2). (D) Comparison of the actin filament elongation rates in the presence of untreated or PKA phosphorylated WT hVASP and the hEVH2 domain alone. Inset shows SDS–PAGE of Coomassie Blue-stained non-phosphorylated and PKA-phosphorylated hVASP. (E) Profilin does not accelerate VASP-mediated actin assembly. The elongation rates of actin and either 200 nM hVASP (left) or 200 nM DdVASP (right) in the presence of 2 μM (grey bars) or 5 μM (black bars) of the indicated profilin isoforms are shown. (F) Profilin–PRD interaction is not required for VASP-mediated filament elongation. The elongation rate of 200 nM DdVASP was not affected by addition of 5 μM profilin Y6D mutant. Data in C–F correspond to means±s.e.m.