Figure 3.
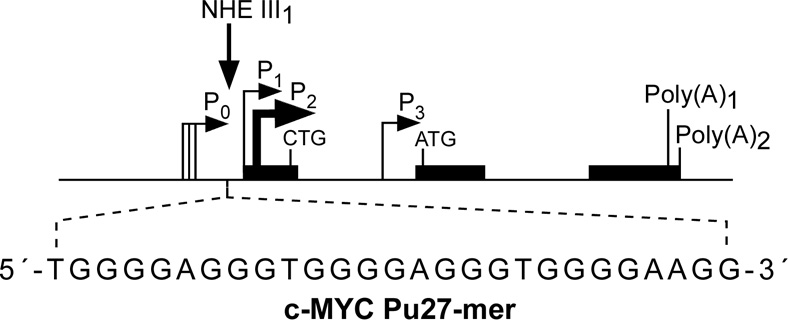
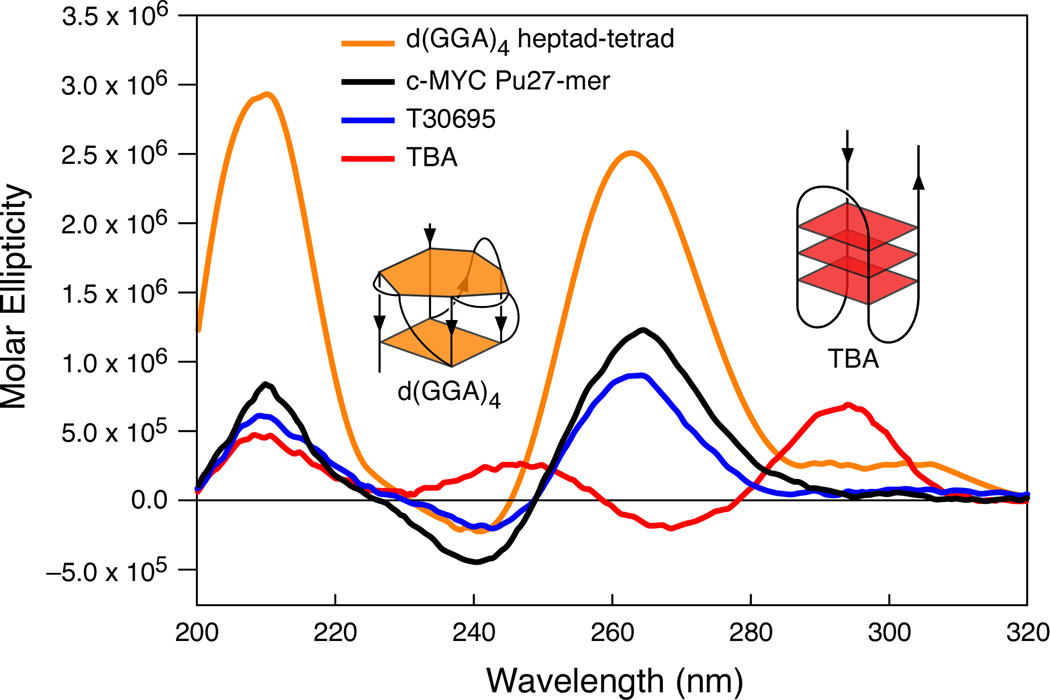
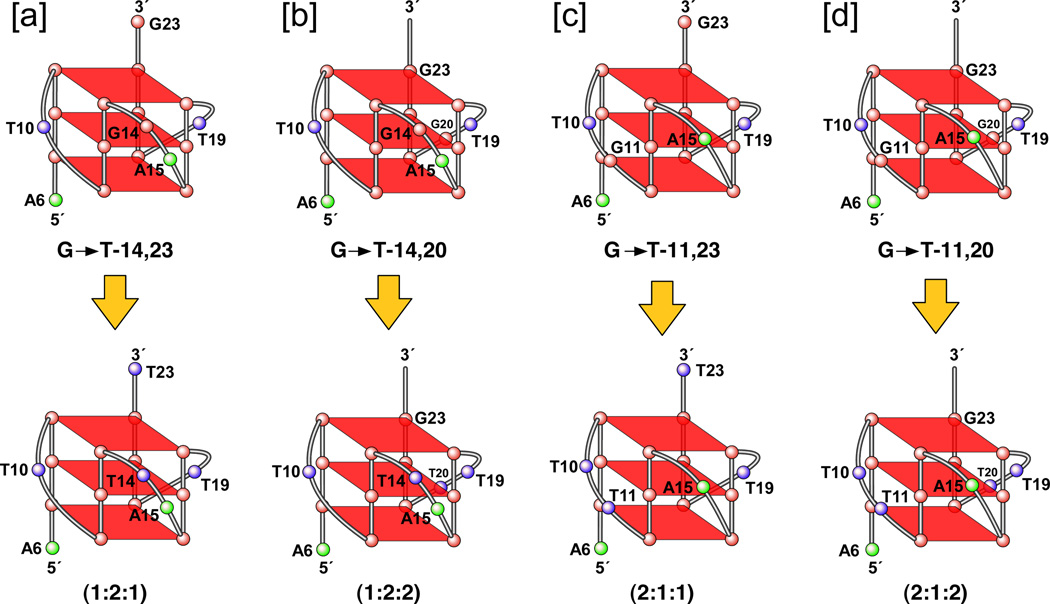
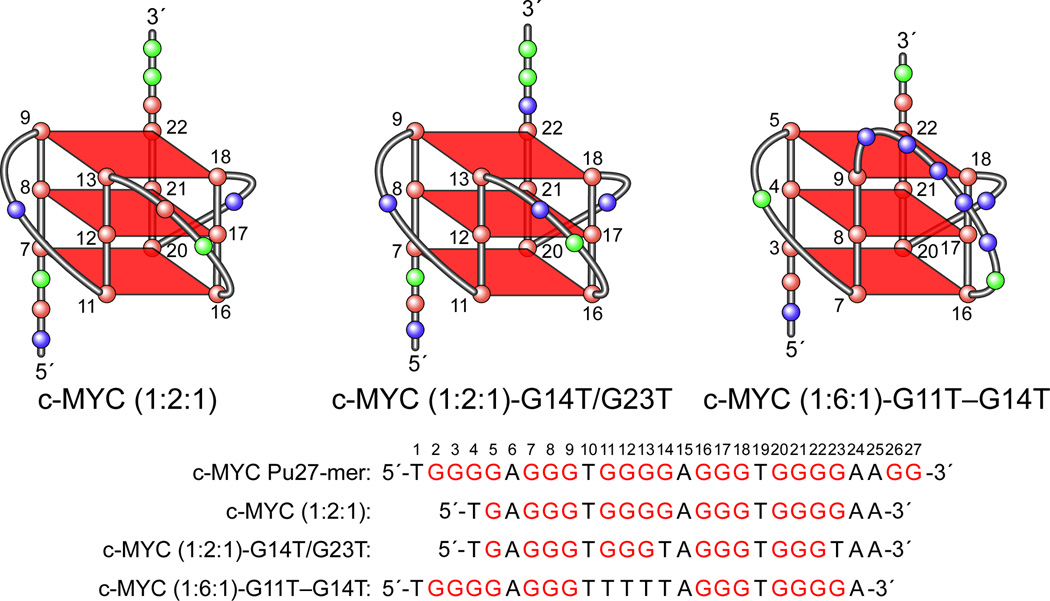
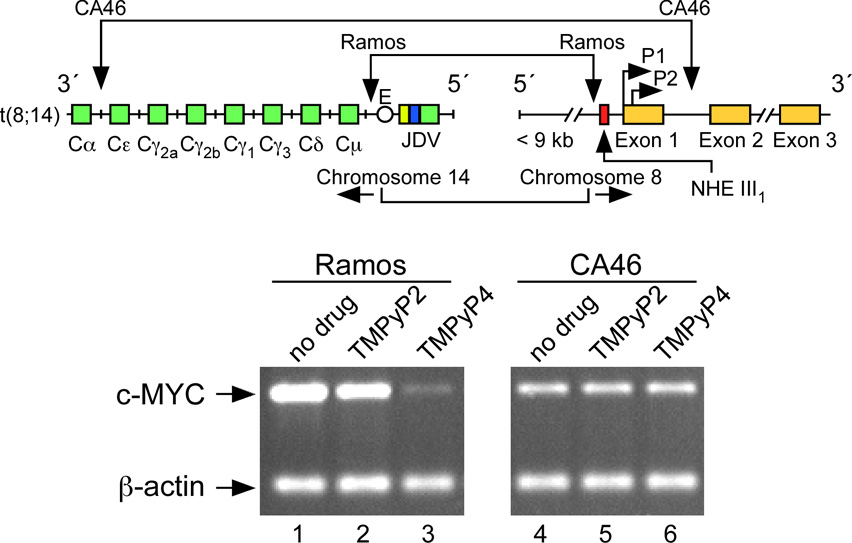
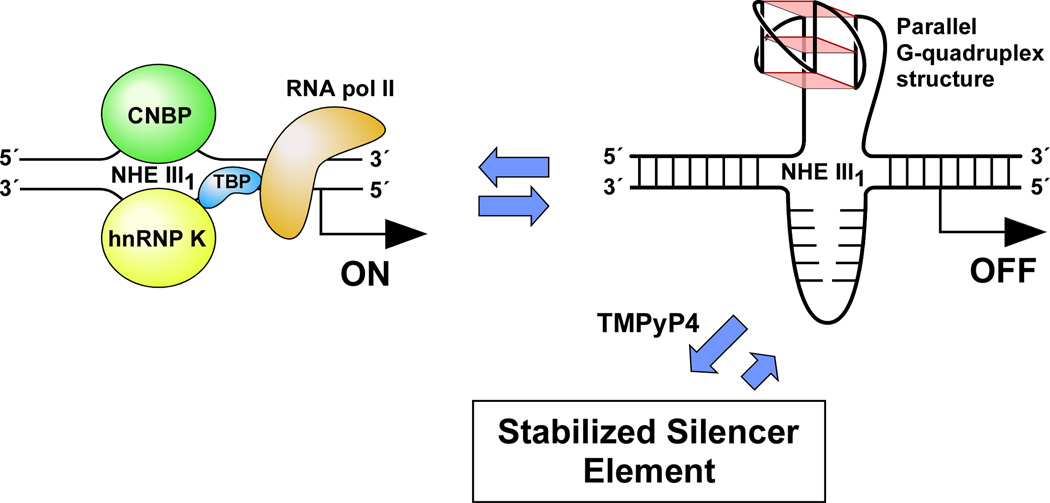
(A) Promoter structure of the c-MYC gene, showing the c-MYC Pu27-mer sequence of the guanine-rich strand upstream of the P1 promoter [41]. (B) CD spectra of the d(GGA)4 oligonucleotide, the c-MYC Pu27-mer, the T30695 oligonucleotide, and the thrombin binding aptamer (TBA). (C) Proposed structures of the four different c-MYC G-quadruplex loop isomers, in which dual G-to-T mutations at positions 11, 14, 20, and 23 result in defined loop isomers. The upper row shows the four proposed isomers and the lower row shows the results of dual G-to-T mutations. In this and subsequent figures, guanines = red, cytosines = yellow, thymines = blue, and adenines = green. (D) Schematic structures of c-MYC (1:2:1), c-MYC (1:2:1)-G14T/G23T, and c-MYC (1:6:1)-G11T–G14T determined by NMR in K+ solution. (E) Diagram of the rearrangements involved in the Ramos and CA46 Burkitt’s lymphoma cell lines (modified from ref. 72). Downward arrows indicate the breakage and rejoining points between chromosomes 14 and 8 for each translocation. RT-PCR for c-MYC and β-actin in Ramos (lanes 1–3) and CA46 (lanes 4–6) cell lines after no treatment (lanes 1 and 4) and treatment with 100 µM TMPyP2 (lanes 2 and 5) and TMPyP4 (lanes 3 and 6) for 48 hr. (F) Model for the activation and repression of c-MYC gene transcription involving the conversion of the paranemic secondary DNA structures (gene off) to purine and pyrimidine single-stranded DNA forms for transcriptional activation. hnRNP K and CNBP are single-stranded DNA binding proteins involved in transcriptional activation. Interaction of the parallel G-quadruplex structure with TMPyP4 stabilizes the gene-off form by conversion to the proposed double-loop G-quadruplex structure that stabilizes the silencer element and results in transcriptional repression [42].