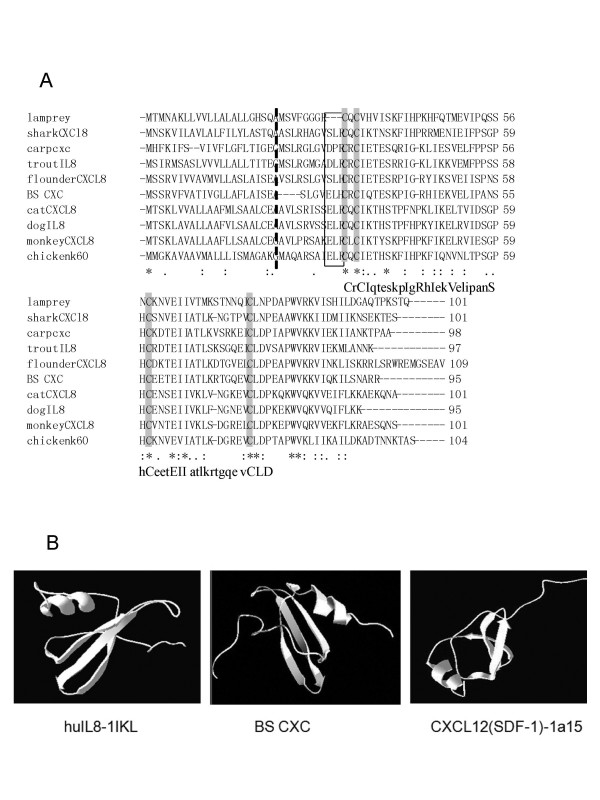
Figure 2.
Amino acid sequence alignment and Comparison of tertiary structure of BS CXC. A. Amino acid sequence alignment of BS CXC with other known CXCL8. The alignment was performed with the Clustal W program. Identical and similar sites are shown with asterisks (*) and dots (. or :), respectively; the broken line indicates the signal peptide cut site. The conserved cysteines are highlighted, and the ELR motif associated with attracting neutrophils in mammals is in the box. B. Comparison of tertiary structure among the human IL-8 model pdb 1Ikl (left), BS CXC (middle) and the CXCL12 (SDF-1) model pdb 1a15. The BS CXC models were predicted by the SWISS-MODEL and Swiss-PdbView software.