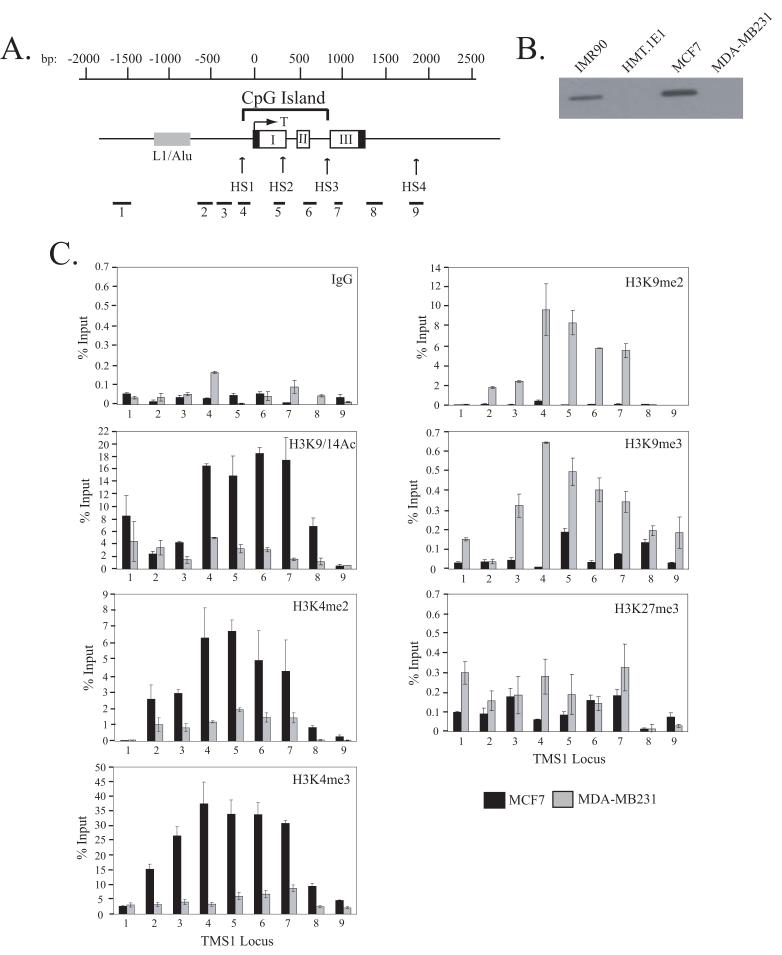
Figure 1.
A) Schematic of the TMS1 genomic locus. The TMS1 gene consists of three exons (I, II and III). Non-coding regions are indicated by black boxes. The nucleotide positions are numbered with respect to the transcription start site (T) and are shown above the gene. The location of the CpG island is marked and spans from approximately -100 to +900 bp. The positions of the hypersensitive sites (HS1-HS4) and an upstream repeat element (L1/Alu) are shown. Primer sets used (1-9) for real-time PCR in ChIP assays are shown below the gene. B) Expression of TMS1. Protein lysates prepared from MCF7, MDA-MB231, HMT.1E1 and IMR90 cells were subject to western blot analysis with antibody against TMS1. C) Distribution of histone H3 modifications across the TMS1 locus. MCF7 and MDA-MB231 cell lines were subjected to ChIP with antibodies against the indicated histone modifications or a rabbit IgG (IgG) control. Immunoprecipitated DNA was amplified by real-time PCR with primer sets indicated in (A). Percent (%) input was determined as the amount of immunoprecipiated DNA relative to input DNA. Each ChIP was repeated at least three times, and although the immunoprecipitation efficiency varied between experiments, the profile of enrichment across the locus was consistent. Shown are the mean +/- standard deviation of triplicate determinations from a representative experiment.