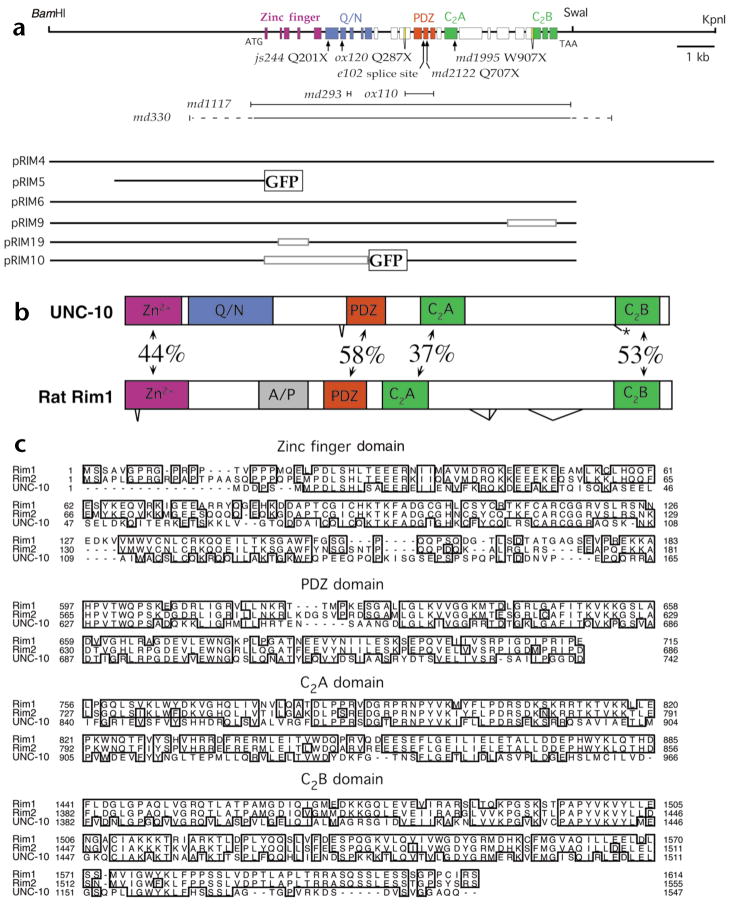
Fig. 1.
Organization of the C. elegans unc-10 gene. (a) Genomic region of the unc-10 gene including selected restriction sites. Exons are shown as colored boxes that define domains using the same color code as in (b). Zinc finger, PDZ and C2 domains were identified using the SMART program43. The glutamine- and asparagine-rich domain (Q/N) was delineated by visual inspection. The position of unc-10 point mutations (arrows) and deletions (|\#x02014;|) are depicted below the genomic map. Genomic regions included in plasmids used in this work are also illustrated. Open boxes in the plasmids indicate deletions of the corresponding unc-10 genomic regions. Green fluorescent protein (GFP) sequences inserted in plasmids are denoted as labeled boxes. (b) Comparison of the domain organization of the C. elegans UNC-10 and rat Rim1 proteins. The percentage sequence identity between shared domains and the positions of alternative splicing are depicted. The alanine and proline rich domain (A/P) and alternative splice forms of Rim1 were identified previously5. (c) Sequence alignments of selected domains of Rim homologs. Alignments were created and formatted using the clustalW and SeqVu programs.