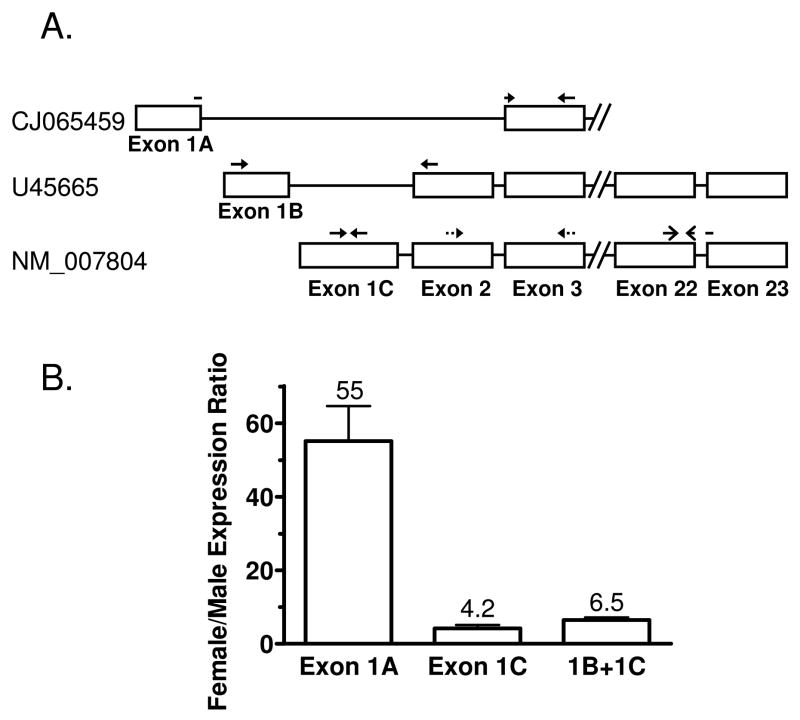
Figure 1. Expression of Cutl2 transcript variants in mouse liver.
Panel A: schematic representation of three different mouse Cutl2 RNAs and qPCR primer pairs used to determine their expression. Cutl2 transcript variants, based on the February 2006 build of the mouse genome, are identified by their GenBank accession numbers. Exon/intron structure shown is not drawn to scale; exons 1A and 1B are separated by 165 nt and exons 1B and 1C by 1951 nts. Exons 1–3 are non-coding. Exon 2 is absent from the exon 1A-containing RNA. The forward primer for this transcript (CJ065459) traverses the exon 1A/exon 3 junction, as indicated. The primer pair amplifying portions of exons 22 and 23 does not discriminate between the three Cutl2 RNA transcripts and was used to assay mouse Cutl2 in a majority of the experiments presented in this study. Panel B: female/male expression ratios of individual Cutl2 transcripts in ICR mouse liver. The ratios were determined for transcripts containing exon 1A, exon 1C and for the exon 1B + exon 1C transcripts together (‘1B + 1C’), as indicated. Relative RNA levels were determined by qPCR analysis of liver RNA from wild-type male and female mice (n=5 per group). The expression levels of each RNA transcript were normalized to the 18S rRNA content of each liver and then calculated relative to the average expression of the corresponding RNA transcript in males. Data shown are mean ± SE. The calculated mean ratios are indicated above each bar. Levels of each Cutl2 transcript showed statistically significant differences between females and males (Student’s t-test, p<0.01).