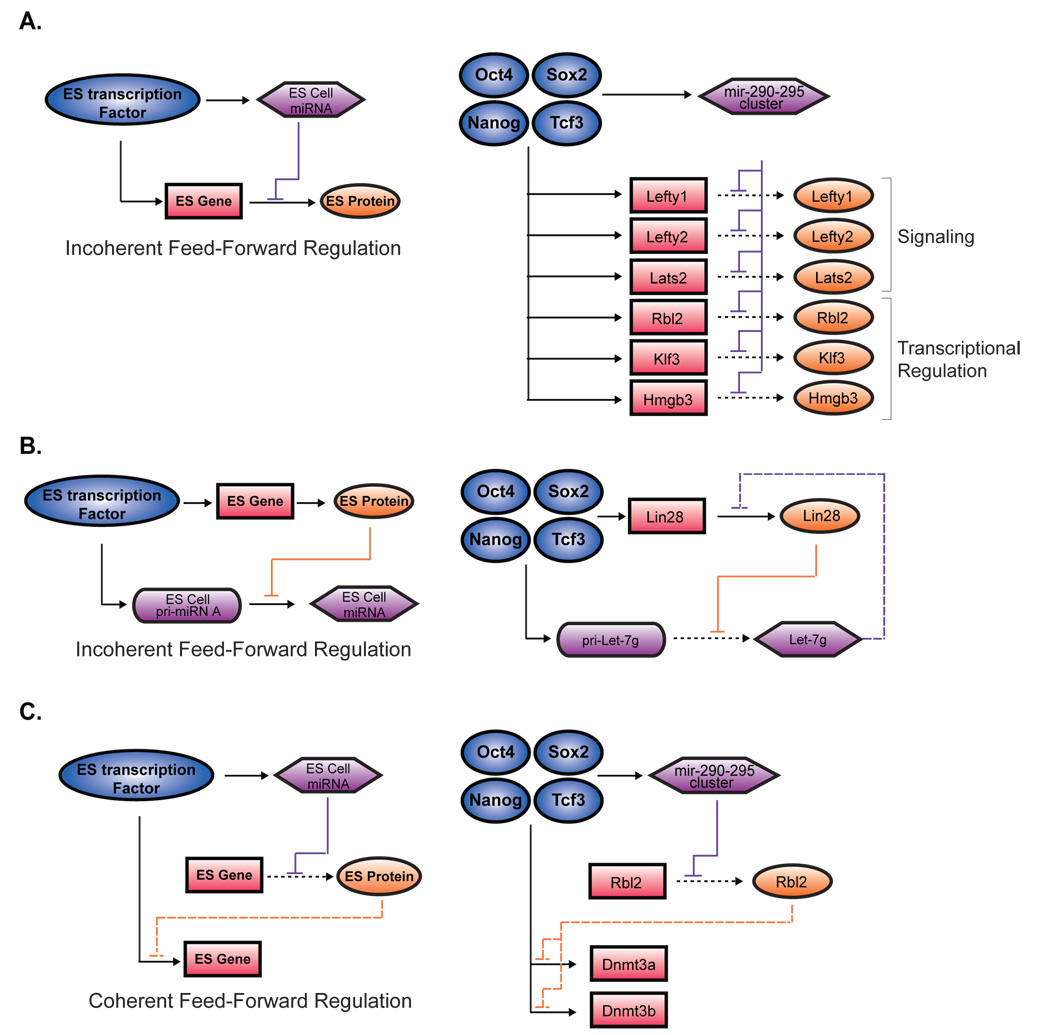
Figure 6. miRNA modulation of the gene regulatory network in ES cells.
A. An incoherent feed-forward motif (Alon, 2007) involving a miRNA repression of a transcription factor target gene is illustrated (left). Transcription factors are represented by dark blue circles, miRNAs in purple hexagons, protein-coding gene in pink rectangles and proteins in orange ovals. Selected instances of this network motif identified in ES cells based on data from Sinkkonen et al., 2008 or data in Figure S11 are shown (right). B. Second model of incoherent feed-forward motif (Alon 2007) involving protein repression of a miRNA is illustrated (left). In ES cells, Lin28 blocks the maturation of primary Let-7g (Visiwanthan et al., 2008). Lin28 and the Let-7g gene are occupied by Oct4/Sox2/Nanog/Tcf3. Targetscan prediction (Grimson et al., 2007), of Lin28 by mature Let-7g is noted (purple dashed line, right). C. A coherent feed-forward motif (Alon 2007) involving miRNA repression of a transcriptional repressor that regulates a transcription factor target gene is illustrated (left). This motif is found in ES cells, where mir-290-295 miRNAs repress Rbl2 indirectly maintaining the expression of Dnmt3a and Dnmt3a, which are also occupied at their promoters by Oct4/Sox2/Nanog/Tcf3 (right).