Fig. 3.
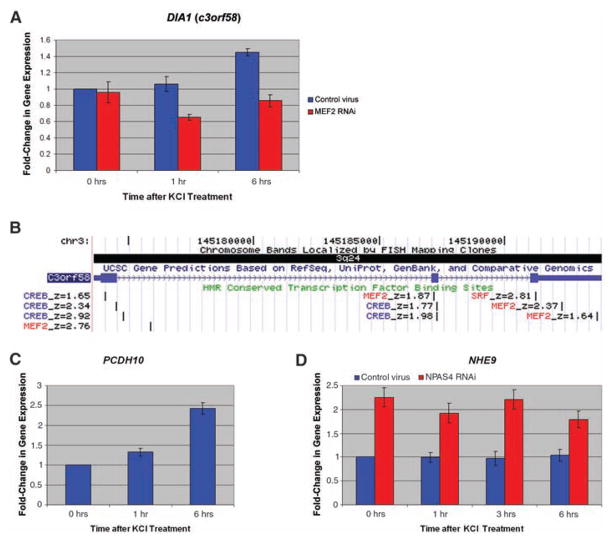
Genes within or juxtaposed to homozygous deletions show activity-dependent gene regulation or are targets of transcription factors regulated by neuronal activity. (A) Activation of DIA1 (c3orf58) gene expression in rat hippocampal cultures (0, 1, and 6 hours) after membrane depolarization with KCl. Control lentivirus shown in blue, and cultures transduced with MEF2A and MEF2D RNAi lentivirus shown in red. (B) The genomic structure of DIA1 (c3orf58), also showing highly conserved transcription factor binding sites based on meeting computation thresholds of conservation in human/mouse/rat alignment with the Transfac Matrix Database (v7.0) (www.gene-regulation.com). Prominent activity-regulated transcription factor sites, namely MEF2/SRF (red) and CREB (blue), are shown. Z-scores of evolutionary conservation are also shown, z-score > 1.64 corresponding to P < 0.05, and z-score > 2.33 corresponding to P < 0.01. (C) Activation of PCDH10 gene expression in rat hippocampal cultures (0, 1, and 6 hours) after membrane depolarization. (D) Activation of NHE9 gene expression in hippocampal cultures (0, 1, 3, and 6 hours) after membrane depolarization with KCl in control lentivirus cultures (blue) and NPAS4 RNAi lentivirus (red).