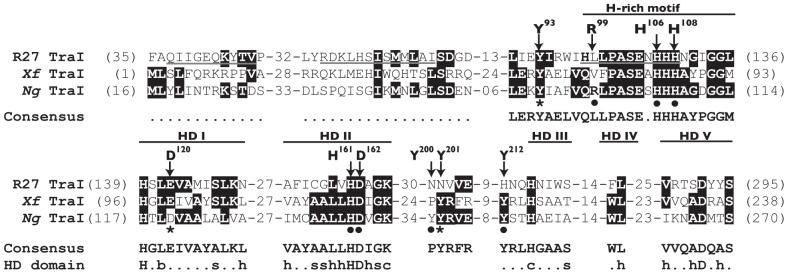
Fig. 2.
Amino acid sequence alignment of predicted relaxases from plasmid R27 (R27 TraI), X. fastidiosa (Xf TraI), and N. gonorrhoeae (Ng TraI). Identical amino acids are highlighted. Underlined sequences in R27 TraI were reported to correspond to the three motifs of classical relaxases. However, the first two motifs are not conserved in the Ng TraI family. The hallmarks of the Ng TraI family are a conserved tyrosine (Tyr93 in Ng TraI) closely followed by a histidine (H)-rich motif, and the HD phosphohydrolase domain. The signature sequence of the H-rich motif is h(Q/H)xhPASExHHHx3GG(L/M)h, where h is a hydrophobic residue and x is any residue. The HD domain motifs are labeled HD I to V (where HD II is the signature motif). The consensus amino acid sequence for the fifty-four proteins in the Ng TraI family of predicted relaxases is shown (the dotted lines represent no consensus). The consensus amino acid sequence for the HD domain is also shown [(b) big, (s) small, (h) hydrophobic, (c) charged, and the capital letters represent invariant amino acids]. The arrows indicate the amino acids in Ng TraI that were the targets for site-directed mutagenesis in this study. Mutations that decrease DNA secretion are indicated with a star. Mutations that have no effect on DNA secretion are indicated with a solid circle.