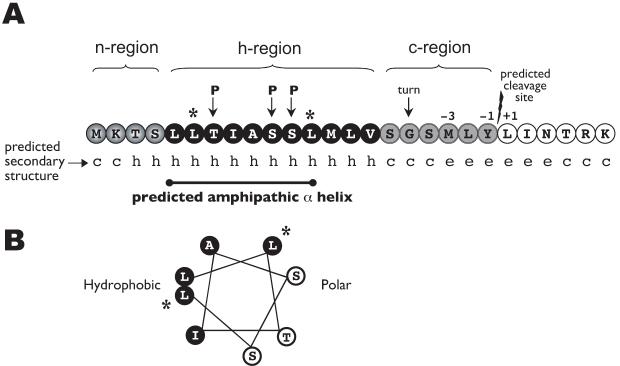
Fig. 4.
Map of the TraI N-terminal region.
A. A SignalP 3.0 algorithm analysis of the Ng TraI amino acid sequence predicted the presence of a signal sequence (0.923 probability) with a cleavage site between Tyr21 and Leu22 (0.5 probability). The hydrophobic (h) region is interrupted by polar residues (P), which results in the secondary structure prediction of an amphipathic α helix (c, coil; h, helix; e, sheet).
B. Helical wheel of amino acids 5 to 12. Black font, white background represents amino acids in the polar side of the helix. White font, black background represents amino acids in the hydrophobic side. *Charged residues were introduced into the hydrophobic side of the predicted amphipathic α helix at positions 6 and 12 (L6K/L12K).