Fig. 9.
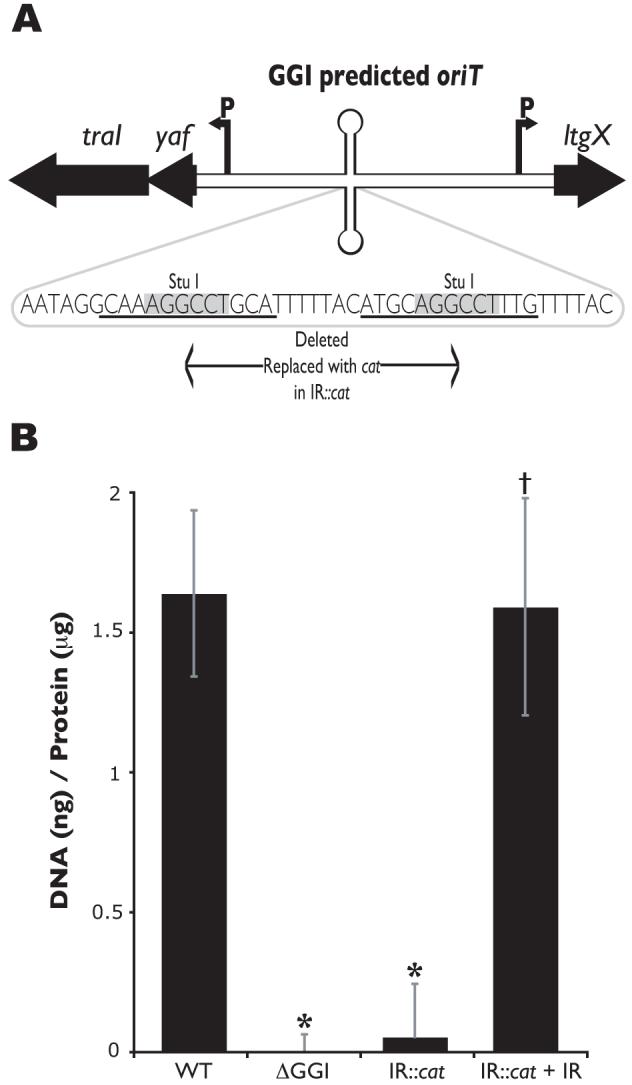
Predicted oriT sequence located in the GGI.
A. Map of the GGI region predicted to contain the oriT. traI encodes the putative relaxase, yaf encodes an unknown protein, and ltgX encodes a putative lytic transglycosylase. Arrows labeled “P” mark the promoter region for the transcripts. The sequence of the yaf-ltgX inverted repeat is shown. An insertion containing a cat marker was introduced at the StuI site of the inverted repeat, deleting the sequence in between the StuI sites.
B. Fluorometric detection of secreted DNA. Piliated gonococcal strains were grown for 2.5h in liquid culture. Cell-free culture supernatants were collected and DNA was detected with the fluorescent DNA-binding dye PicoGreen and normalized to total protein in the cell pellet. MS11 was used as the wild-type (WT) strain and ND500 (ΔGGI in MS11) as the negative control. The results are an average of at least four independent experiments. *p-value<0.003 when compared to wild-type. †p-value<0.003 when compared to its respective mutant.
