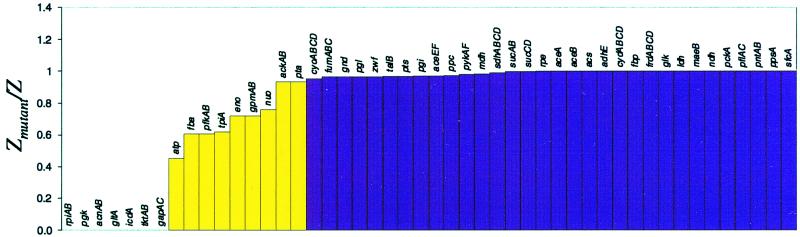
Figure 2.
Gene deletions in E. coli MG1655 central intermediary metabolism; maximal biomass yields on glucose for all possible single gene deletions in the central metabolic pathways. The optimal value of the mutant objective function (Zmutant) compared with the “wild-type” objective function (Z), where Z is defined in Eq. 3. The ratio of optimal growth yields (Zmutant/Z). The results were generated in a simulated aerobic environment with glucose as the carbon source. The transport fluxes were constrained as follows: βglucose = 10 mmol/g-dry weight (DW) per h; βoxygen = 15 mmol/g-DW per h. The maximal yields were calculated by using FBA with the objective of maximizing growth. The biomass yields are normalized with respect to the results for the full metabolic genotype. The yellow bars represent gene deletions that reduced the maximal biomass yield to less than 95% of the in silico wild type.