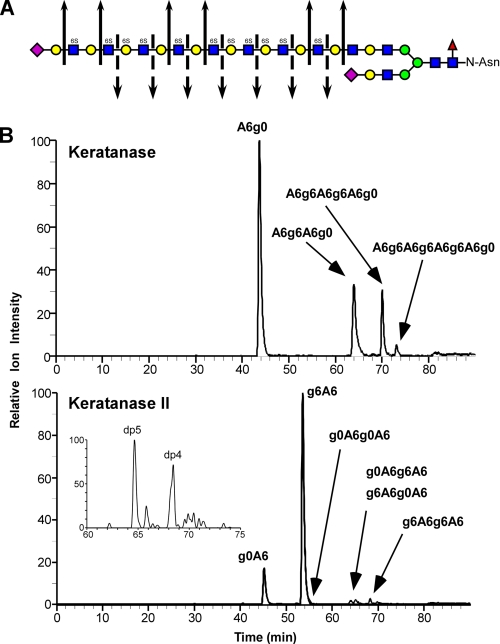
FIGURE 4.
Qualitative structural analysis of KS GAG chains using GRIL-LC/MS. A, generalized structure of corneal KS I showing the cleavage sites for keratanase (solid arrows) and keratanase II (broken arrows). The symbols used for monosaccharides subunits: sialic acid (purple diamond), galactose (yellow circle), N-acetylglucosamine (blue squares), mannose (green circles), and fucose (red triangle) are as previously described (grtc.ucsd.edu/symbol.html). The presence of 6-O-sulfate is also indicated (6S) above each sulfated monosaccharide. B, bovine corneal KS was digested with either keratanase (“Experimental Procedures”), aniline-tagged and subjected to LC/MS for analysis of digestion products. The chromatographs show the relative ion intensity (XIC) of digestion products eluting from the C18 column. The indicated peak assignments were based on retention time, mass, and the known activity of the keratanases used. The inset in the lower panel shows the elution of two species released by keratanase II digestion with degree of polymerization of 4 and 5 (dp4 and dp5) based on mass. The XIC traces were generated with the appropriate m/z values for KS disaccharides listed in Table 1 and for m/z values for the major expected ions of oligosaccharides released after keratanase digestion. For keratanase, these included: dp4 ([M-3H+DBA]-2, m/z = 596), dp6 ([M-5H+3DBA]-2, m/z = 987), and dp8 ([M-5H+3DBA]-2, m/z = 1250). For keratanase II these included dp4 species g0A6g0A6 ([M-2H]-2, m/z = 491.5), g0A6g6A6 and g6A6g0A6 ([M-3H+DBA]-2, m/z = 596), and g6A6g6A6 ([M-4H+2DBA]-2, m/z = 700.5). The inset XIC trace in the Keratanase II panel was generated for the putative dp5 species Neu5Ac-g0A6g0A6 ([M-2H]-2, m/z = 637), which also reveals a putative minor adduction ion for g6A6g6A6 ([M-3H+DBA]-2, m/z = 636). The mass spectra of the small peak eluting just after 80 min in the upper chromatogram and the small peak eluting after the putative g6A6g6A6 peak in the lower chromatogram do not correspond to any known keratan sulfate-derived oligosaccharides and are most likely contaminants.