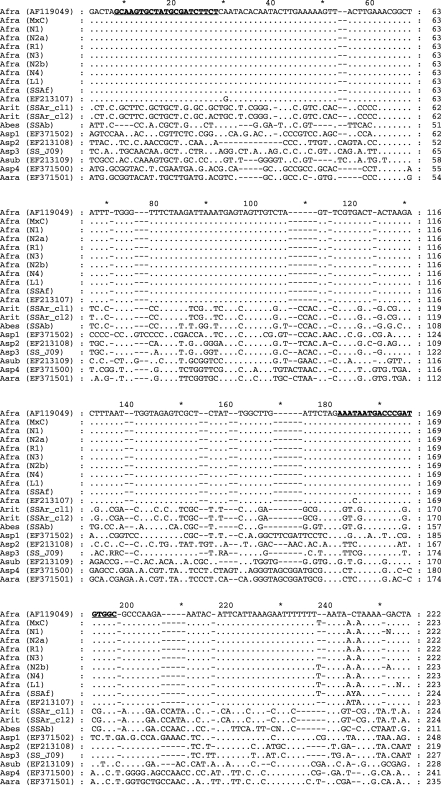
Fig. 1.
Nucleotide sequence alignment of partial fragment of the ITSl-rRNA gene of Aphelenchoides fragariae, GenBank Accession AFl 19049 with A. fragariae isolates (Afra) from monoxenic culture (MxC); from naturally infected host plants from six different nursery (N), retail (R) or landscape (L) locations in North Carolina including × Heucherella ‘Sunspot’ (Nl) (EU219917), Dryopteris remota (N2a), Abelia × grandiflora ‘Edward Goucher’ (Rl), Lantana camara ‘Miss Huff’ (N3) (EU219918), Arachnoides simplicior ‘Variegata’ (N2b), Buddleja davidii ‘Petite Plum’ (N4) (EU219919), Hosta sp. (LI); A. fragariae isolate, Pteris longifolia, Moscow, Russia (SSaf) (EU186070); A. fragariae isolate, Aconitum napellus, Netherlands (EF213107); A. ritzemabosi isolate with two ITS clones, Sambucus racemosa, Moscow, Russia (SSAr_cll and SSAr_cl2)(EU186067, EU186068); A. besseyi isolate, rice, Krasnodar region, Russia (SSAb)(EU186069); Aphelenchoides sp. 1, tulip, Netherlands (Aspl) (EF371502); Aphelenchoides sp. 2, tulip, originally identified as A. ritzemabosi, Netherlands (Asp2) (EF213108); Aphelenchoides sp. 3, University of California-Riverside collection (SS-J09) (EU186071); A. subtenuis, Crocus vernus, Netherlands (Asub) (EF213109); Aphelenchoides sp. 4, Narcissus, originally identified as A. besseyi, Netherlands (Asp4) (EF371500); A. arachidis, Sternbergia, Netherlands (Aara) (EF371501). Aphelenchoides fragariae-speciiic primers (AfragFl and AfragRl) are in bold type and underlined. Identities with the A. fragariae consensus sequence are indicated by a period; gaps introduced to maximize alignment are marked by hyphens.