Abstract
Sexual dimorphism in mammalian liver impacts genes affecting hepatic physiology, including inflammatory responses, diseased states and the metabolism of steroids and foreign compounds. Liver sex-specificity is dictated by sex differences in pituitary growth hormone (GH) secretion, with the transcription factor STAT5b required for intracellular signaling initiated by the pulsatile, male plasma GH profile. STAT5a, a highly homologous but minor liver STAT5 form, also responds to sexually dimorphic plasma GH stimulation, but is unable to compensate for the loss of STAT5b and the associated loss of sex-specific liver gene expression. A large-scale gene expression study was conducted using 23,574-feature oligonucleotide microarrays and livers of male and female mice, both wild-type and Stat5a-inactivated, to elucidate any dependence of liver gene expression on STAT5a. Significant sex differences in expression were found for 2482 mouse genes, 1045 showing higher expression in males and 1437 showing higher expression in females. In contrast to the widespread effects of the loss of STAT5b, STAT5a deficiency had a limited but well defined impact on liver sex-specificity, with 219 of 1437 female-predominant genes (15%) specifically decreased in expression in STAT5a-deficient female liver. Analysis of liver RNAs from wild-type mice representing three mixed or outbred strains identified 1028 sexually dimorphic genes across the strains, including 405 female-predominant genes, of which 91 (23%) required STAT5a for normal expression in female liver. These findings highlight the importance of STAT5a for regulation of sex-specific hepatic genes specifically in female liver, in striking contrast to STAT5b, whose major effects are restricted to male liver.
Keywords: STAT5a, microarray, liver sexual dimorphism, growth hormone action, strain-dependent gene expression
Introduction
Growth hormone (GH) regulates gene expression in several tissues, most notably liver. GH is secreted by the pituitary gland in a sex-dependent manner under the regulation of gonadal steroids (12, 26). This, in turn, leads to substantial sex differences in GH-regulated liver gene expression (17, 31). In the rat, plasma GH levels are highly pulsatile in males, where hormone peaks every ~3.5 hr are followed by a GH-free interval lasting ~2 hr, whereas in females, GH is present in plasma in a more continuous manner (12). Mice also show sexually dimorphic GH secretory patterns, with females characterized by more frequent GH pulses and a distinctly shorter GH-free interpulse interval than males (16). These sex-dependent plasma GH profiles control liver gene expression at the level of transcription, as demonstrated several GH-regulated genes that encode liver cytochrome P450 (Cyp) enzymes active in the metabolism of steroids, drugs and environmental chemicals (29).
GH signaling is initiated by the binding of GH to its cell surface receptor, which activates JAK2, a GH receptor-associated tyrosine kinase. JAK2, in turn, phosphorylates GH receptor on multiple cytoplasmic domain tyrosine residues, several of which serve as docking sites for multiple STAT (signal transducer and activator of transcription) transcription factors (6, 10), including STAT5a and STAT5b (7, 8, 21). Each STAT is then phosphorylated on a single tyrosine residue, which leads to STAT homo- and heterodimer formation, translocation to the nucleus, binding to STAT5 DNA response elements, and stimulation of gene transcription. Both STAT5a and STAT5b are directly activated in male rat liver in response to each incoming plasma GH pulse (4), whereas in livers of female rats, the persistence of plasma GH stimulation leads to partial desensitization of the STAT5 signaling pathway and substantially lower nuclear STAT5b protein than the peak levels seen in males (3, 4, 30). Based on these and other findings, STAT5b has been proposed to serve as a mediator of the sex-dependent effects that GH has on liver gene expression (27). This proposal is supported by the characterization of STAT5b-deficient male mice, which display a reduced body growth rate at puberty and a loss of sex-specific liver expression of Cyps and other genes (9, 23, 25). Microarray analysis has demonstrated that the impact of STAT5b on sex-dependent liver gene expression is global, with 90% of male-predominant genes down-regulated and 61% of female-predominant genes up-regulated in livers of STAT5b-deficient male mice (5). Studies of STAT5a-deficient mice (15) raise the possibility that STAT5a may be required for expression of certain female-specific mouse Cyp proteins (20), however, this has not been investigated in a systematic way.
Sexual dimorphism of liver gene expression is extensive, affecting at least 1500 genes (5, 32), including many members of the Cyp gene superfamily (29). These sex-differences may be impacted by genetic factors, which are manifest as strain differences in sex-dependent gene expression. For example, certain Cyp2b steroid 16α-hydroxlase enzymes show strain-dependent differences that affect sex-specificity (19). Moreover, Cyp2a4 is subject to strain-specific regulation associated with a recessive mutation that de-represses its expression in males (2). Strain-dependent expression also characterizes sex-specific genes regulated by regulators of sex-limitation (Rsl), transcriptional repressors that are deficient in certain mouse strains and modulate the expression of Slp, Mups and certain other male-specific, GH-regulated genes (14, 24). Mutations in Rsl1 and Rsl2 serve as examples of strain-specific alleles in transcriptional regulators that impact the sex-specificity of GH-responsive genes in the liver (13, 28). It is uncertain whether these or other strain-dependent factors have a widespread effect on the sex-specificity of hepatic gene expression.
The present study uses microarray technology to assess the impact of STAT5a deficiency on the sex specificity of liver gene expression. In addition, global assessment of sex specificity was investigated in three different mixed or outbred mouse strains to identify a robust set of strain-independent, sex-specific genes, as well as sets of genes whose sex-dependence apparently varies with the genetic background. Our findings reveal that Stat5a disruption affects the expression of a discrete subset of sex-dependent genes in female mouse liver, in sharp contrast to Stat5b disruption, which selectively abrogates sex-specific gene expression in male liver.
Materials and Methods
Animals
STAT5a-deficient mice were prepared by targeted disruption of the Stat5a gene (15). Livers were collected from 6–9 wk old male and female 129J × Black/Swiss mice, wild-type and STAT5a-knockout (KO). Livers were also collected from 8–10 wk old male and female wild-type mice of the ICR strain (Taconic, Inc., Germantown, NY). Livers were snap frozen in liquid nitrogen and stored at −80°C until use.
RNA isolation
Total RNA was isolated from ~0.1 g frozen mouse liver using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. Liver RNAs prepared from 12 individual 129J × Black Swiss mice (30 μg RNA per liver, dissolved in DEPC-treated water) were used in the present study: 3 wild-type males, 3 STAT5a-KO males, 3 wild-type females, and 3 STAT5a-KO females. Ten individual ICR mouse liver RNAs (5 wild-type female livers and 5 wild-type male livers), kindly provided by Dr. Ekaterina Laz of this laboratory, were used to prepare 2 pooled liver RNA samples (n = 2 livers and n = 3 livers, respectively). Two of the individual livers for each sex were also used to prepare single liver RNA samples.
qPCR analysis
Liver RNA samples were converted to cDNA and used in qPCR assays for individual genes from groups 13 and 14 (Table 3) using methods detailed elsewhere (9). Amplification of a single, specific product during qPCR cycling was verified by examination of dissociation curves of each amplicon. Relative RNA levels were determined after normalization to the 18S RNA content of each sample. Statistical analysis was carried out by Student’s t-test using GraphPad Prism software version 4 (San Diego, CA). P-values less than 0.05 were considered significant. qPCR primer design was carried out using Primer Express software (Applied Biosystems), and all primers were verified with respect to their specificity for the target transcript by BLAT analysis of the mouse genome (February 2006 assembly) at http://genome.ucsc.edu/cgi-bin/hgBlat. Primer sequences are shown in Table S1.
Table 3. Distribution of 3905 differentially expressed genes from the present STAT5a –KO study and 2231 differentially expressed genes from the STAT5b-KO study (5) within co-expressed gene groups.
Genes are grouped based on the Total Flagging Sum (TFS) system (Materials and Methods). This is indicated by the direction of response for each ratio as detailed in the four rightmost columns as indicated by male (M), female (F), and Up or Down-regulation in wild-type and STAT5a- or STAT5b-knockout males or females, or no response (−), as indicated. Twenty-eight gene groups, each containing at least 15 genes in one of the two data sets, were assigned group numbers. 55 genes from the STAT5b-KO data set, representing less than 2.5% of the genes, were distributed among 20 additional gene groups and are classified as Other. 85 genes from the STAT5a-KO data set, representing ~ 2% of the genes, were among 18 additional gene groups, which were also classified as Other. Percentages are based upon the 3905 and 2231 genes
| Group | Total Flagging Sum (TFS) | STAT5a | STAT5a (%) | STAT5b | STAT5b (%) | Sex Specificity (WT) | Sex Specificity (KO) | Response to the loss of STAT5a in males | Response to the loss of STAT5a in females |
|---|---|---|---|---|---|---|---|---|---|
| 1A | 3.2200 | 27 | 0.7 | 560 | 25.1 | M | - | Down | - |
| 1B | 3.1100 | 26 | 0.7 | 357 | 16.0 | F | - | Up | - |
| 2A | 15.2211 | 6 | 0.2 | 44 | 2.0 | M | M | Down | Down |
| 2B | 15.1122 | 3 | 0.1 | 39 | 1.7 | F | F | Up | Up |
| 3A | 11.2201 | 25 | 0.6 | 139 | 6.2 | M | M | Down | - |
| 3B | 11.1102 | 18 | 0.5 | 36 | 1.6 | F | F | Up | - |
| 4A | 1.2000 | 123 | 3.1 | 58 | 2.6 | M | - | - | - |
| 4B | 1.1000 | 280 | 7.2 | 229 | 10.3 | F | - | - | - |
| 5A | 9.2001 | 783 | 20.1 | 20 | 0.9 | M | M | - | - |
| 5B | 9.1002 | 851 | 21.8 | 43 | 1.9 | F | F | - | - |
| 6A | 7.2210 | 0 | 0.0 | 15 | 0.7 | M | - | Down | Down |
| 6B | 7.1120 | 0 | 0.0 | 15 | 0.7 | F | - | Up | Up |
| 7A | 8.0001 | 353 | 9.0 | 39 | 1.7 | - | M | - | - |
| 7B | 8.0002 | 585 | 15.0 | 127 | 5.7 | - | F | - | - |
| 8A | 2.0200 | 61 | 1.6 | 86 | 3.9 | - | - | Down | - |
| 8B | 2.0100 | 24 | 0.6 | 133 | 6.0 | - | - | Up | - |
| 9A | 4.0010 | 83 | 2.1 | 30 | 1.3 | - | - | - | Down |
| 9B | 4.0020 | 88 | 2.3 | 17 | 0.8 | - | - | - | Up |
| 10A | 6.0210 | 16 | 0.4 | 36 | 1.6 | - | - | Down | Down |
| 10B | 6.0120 | 14 | 0.4 | 21 | 0.9 | - | - | Up | Up |
| 11A | 10.0101 | 36 | 0.9 | 51 | 2.3 | - | M | Up | - |
| 11B | 10.0202 | 37 | 0.9 | 13 | 0.6 | - | F | Down | - |
| 12A | 12.0022 | 71 | 1.8 | 40 | 1.8 | - | F | - | Up |
| 12B | 12.0011 | 41 | 1.0 | 11 | 0.5 | - | M | - | Down |
| 13A | 5.2020 | 26 | 0.7 | 2 | 0.1 | M | - | - | Up |
| 13B | 5.1010 | 164 | 4.2 | 14 | 0.6 | F | - | - | Down |
| 14A | 13.2021 | 28 | 0.7 | 0 | 0.0 | M | M | - | Up |
| 14B | 13.1012 | 51 | 1.3 | 1 | 0.0 | F | F | - | Down |
| Other | Other | 85 | 2.2 | 55 | 2.5 | M/F | M/F | Up/Down | Up/Down |
Microarray analysis
Global expression analysis was determined using the 23,574-feature mouse Rosetta/Merck Mouse TOE 75k Array 1 (GEO Platform: GPL 3562; Agilent Tech., Palo Alto, CA), with each feature corresponding to a single 60-mer oligonucleotide anti-sense probe. Each probe is herein referred to as representing a distinct gene, although the actual number of genes analyzed is likely to be smaller than this number due to the presence of non-annotated sequences, some of which may duplicate results for other genes represented on the chip. Liver RNA samples (n = 3 independent biological replicates for each of 4 sex-genotype combinations) were used in 4 separate competitive hybridization experiments in a loop design: male wild-type vs. female wild-type (M-WT:F-WT); male wild-type vs. male STAT5a-KO (M-WT:M-KO); female wild-type vs. female STAT5a-KO (F-WT:F-KO); and male STAT5a-KO vs. female STAT5a-KO (M-KO:F-KO). Each RNA sample was labeled with Cy3-dUTP or Cy5-dUTP in a reverse transcription reaction to generate fluorescent-labeled cDNA. The Cy3-labeled cDNA from one of the three male wild-type samples was mixed with the Cy5-labeled cDNA from one of the three female wild-type samples. The opposite-labeled cDNA from both samples were also mixed. Together, these two mixed cDNA samples are considered a fluorescent reverse pair (‘dye swap’) and were prepared for each of the four sets of hybridization experiments. Two microarrays, one for each mixed cDNA sample, were hybridized for each fluorescent reverse pair. Three fluorescent reverse pairs, corresponding to the three samples of each liver RNA, were hybridized for each of the four microarray comparisons, giving a total of 24 microarrays for the STAT5a data set. Four fluorescent reverse pairs, corresponding to the 2 pooled RNA and 2 single liver RNA samples, were hybridized for the wild-type male and wild-type female comparison of the ICR mice. The fluorescence intensity values obtained from each microarray were normalized using Rosetta Resolver (Rosetta Biosoftware, Seattle, WA). The two halves of a single fluorescent reverse pair were averaged to remove any potential dye-dependent effects on the reported expression ratios. Probes with intensities resulting from over-saturation were removed from consideration. Expression ratios obtained in this study are included in Table S2. The data are also available for query or download from the Gene Expression Omnibus (GEO) web site at NCBI (http://www.ncbi.nlm.nih.gov/geo) as GEO series GSE7169 and GSE7170. GenBank accession numbers and associated gene names, gene descriptions and Unigene numbers based on Unigene build 161 were obtained for unassigned features using the BLAST-Like Alignment Tool (BLAT) located on the UCSC Genome Browser web site. Assignments were based on the best matches (identity of ≥ 56 out of 60 nucleotides) to the 60-nucleotide probe sequences of each probe. Probes that returned multiple accession numbers were verified to make sure that all accession numbers returned represented the same gene. Probe sequences are available upon request.
Statistical analysis
Moderated t-statistics, using standard errors moderated by a simple Bayesian model, were generated using the LIMMA (Linear Models for MicroArray data) package as part of the Bioconductor project within the R statistical program (22). This is the same p value used in our prior microarray study on the effect of STAT5b in mouse liver gene expression (5). A filter (p < 0.05) was applied to the p values to determine the statistical significance of each gene’s differential expression for each of the four microarray comparisons (M-WT:F-WT, M-WT:M-KO, F-WT:F-KO, M-KO:F-KO). Multiple testing correction methods, such as Bonferroni or Holm step-down, were not applied to the p values associated with these four comparisons because these options depend heavily on the independence of each gene’s expression and thus filter out many bona fide regulated genes to avoid all type I errors; they are thus too restrictive in their effort to avoid false positives, as noted elsewhere (1). To compare sex-specificity across strains, microarrays comparing wild-type male vs. female gene expression in three distinct genetic backgrounds were averaged (n = 10 arrays) to determine the robustness of sex-specificity across all three strains and experiments. A differential expression filter (mean ratio > 1.5 or < 0.66) was applied to the gene expression values deemed statistically significant by the p < 0.05 filter. Threshold values of > 1.5 or < 0.66 were chosen as described previously (1, 5). Genes that passed the criteria for statistical significance and the differential expression threshold for at least one of the four mean ratios were included in our analyses (3905 genes in total; see Results). 1810 genes from the M-WT:F-WT comparison of ICR strain mice were included in our analyses as well (see Results). Analysis of Variance (ANOVA) with a Benjamini and Hochberg False Discovery Rate of 0.001 was used, as implemented in GeneSpring GX 7.3.1 software (Agilent Technologies, Santa Clara, CA), to determine strain-specificity for the genes that met the threshold criteria for only one of the three independent data sets for M-WT:F-WT comparison.
A system of binary and decimal flags was applied for clustering genes based on expression ratios obtained in all four microarrays, as previously described in our STAT5b-KO microarray study (5). For the purpose of this clustering, threshold ratios for differential expression were reduced to values of > 1.25 and < 0.8 for the three arrays that involved STAT5a-KO liver RNAs, with retention of the p < 0.05 threshold for statistical significance. Average ratios meeting these threshold and significance criteria contributed to the binary- and decimal-based flag. Thus, genes with a M-WT:F-WT microarray ratio meeting the criteria were assigned a binary flag value of 1, while genes meeting the criteria for the M-WT:M-KO, F-KO:F-WT and F-KO:M-KO microarrays were respectively assigned binary flag values of 2, 4 and 8. Genes not meeting these criteria were assigned flag values of 0. The sum of these binary-based flag values defines the whole number portion of the flag, and was used as a simple method to identify which of the four microarrays met our criteria for inclusion for any given gene of interest, regardless of the direction (up or down) of the regulation. The flag value was then extended using decimal values of 0.1, 0.01, 0.001 and 0.0001, or 0.2, 0.02, 0.002 and 0.0002, for each of the four microarrays, to indicate the direction of regulation between the two conditions on the microarray. Thus, average ratios for the M-WT:F-WT microarray > 1.5 were assigned a decimal value of 0.2, to indicate up-regulation, while average ratios < 0.66 were assigned a value of 0.1, to indicate down-regulation. The three other microarray ratios were similarly flagged, based on threshold values of > 1.25 and < 0.8, as indicated above, by advancing to a new decimal position for each microarray (i.e., the M-WT:M-KO flag is in the hundredths position, and so on). For each gene, the resulting binary sum describes which microarray ratios met the selection criteria and the four-digit decimal value describes the direction of regulation (Total Flagging Sum, TFS).
GO (Gene Ontology) term enrichment analysis
GO molecular function annotations were analyzed for term enrichment using the GO Browser within GeneSpring GX software. Because the GO term assigned to each gene represents a sub-class of its parent class within the ontology, each gene was also assigned all parent GO class terms. For each of the GO categories present among each list of genes with GO annotations, the number of genes assigned that term was calculated along with a count of all genes on the microarray assigned that term. The p-value for each category was calculated based on the total number of genes on the microarray within the category and the number of genes in the group being tested within the category. The p-value represents the likelihood that at least as many genes would occur if a list of equal size were selected by chance from the total gene count.
Results
Experimental design
A large-scale expression study was conducted to investigate the role of STAT5a in the sex-specificity of mouse liver gene expression. RNA was isolated from livers of male (M) and female (F) mice that were either wild-type (WT) or contained a targeted disruption of the Stat5a gene (KO). RNA samples representing three biological replicates for each sex-genotype combination were competitively hybridized to 23,574-feature oligonucleotide microarrays. Four sets of competitive hybridizations were carried out: 1) M-WT vs. F-WT; 2) M-WT vs. M-KO; 3) F-WT vs. F-KO; and 4) M-KO vs. F-KO. Normalized hybridization intensities were used to calculate mean expression ratios based n = 3 biological replicates for each sex-genotype combination, and p-values were calculated using the LIMMA toolset within the R statistical program (22). Probes representing 3905 genes (16.6% of the total number of probes) met the threshold criteria for differential expression (average expression ratio >1.5-fold and a significance level of p < 0.05) for at least one of the four sex-genotype combinations. Thus, these 3905 genes were expressed in a sex-specific manner in either wild-type or STAT5a-KO mice or responded to the loss of STAT5a in either males or females. Average expression ratios for the 3905 genes of interest are listed in Table S2.
Overview of sex-specificity and impact of STAT5a deficiency
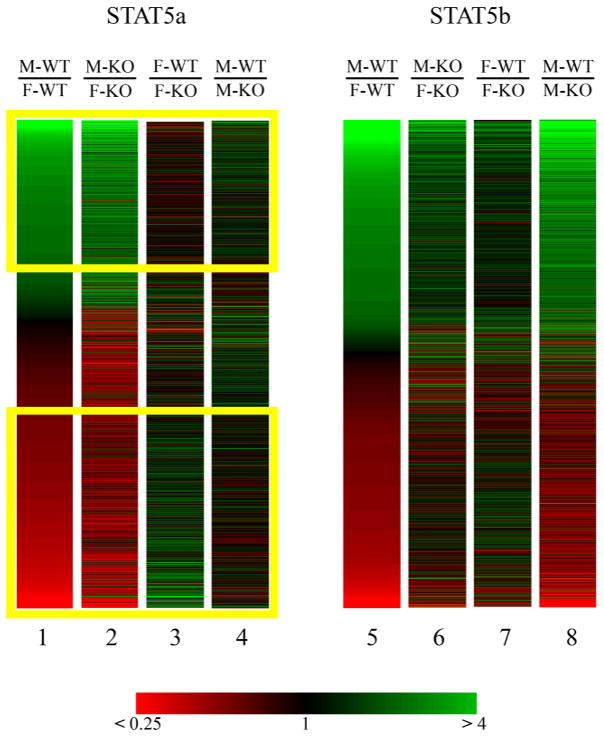
Gene expression differences between wild-type male and wild-type female liver were found for 2482 of the 3905 genes of interest (64%), indicating these genes are sexually dimorphic in wild-type liver. These genes are colored green (M-WT > F-WT) or red (M-WT < F-WT) in Fig. 1, lane 1, where they are displayed at the far ends of a false-color heat map containing all 3905 genes sorted by average M-WT:F-WT ratio. Expression of 1045 of the 2482 genes was male-predominant in wild-type mouse liver (M-WT:F-WT >1.5; lane 1, upper yellow box, Fig. 1), while 1437 genes were female-predominant (M-WT:F-WT <0.667; lane 1, lower yellow box, Fig. 1).
Fig. 1. False color heat maps for expression profiles of 3905 differentially expressed genes from the STAT5a-KO data set and 2231 differentially expressed genes from the STAT5b-KO data set.
Genes are depicted based on their mean expression ratios across four experimental pairings, as indicated on top of each column. Genes are colored according to the colorbar at the bottom, ranging from bright green for an expression ratio >4 to bright red for an expression ratio <0.25, with black corresponding to a ratio of 1. In lanes 1–4, 3905 genes from the present STAT5a data set are sorted according to the mean M-WT:F-WT ratio. The upper and lower yellow boxes enclose genes determined to be male-specific and female-specific, respectively (M-WT:F-WT >1.5 or <0.66 at p<0.05). In lanes 5–8, 2231 genes from the STAT5b-KO data set (5) are sorted according to M-WT:F-WT ratio.
Hierarchical clustering of the four microarray data sets grouped the male-female comparisons for wild-type and knockout mice together (Pearson’s correlation coefficient = 0.726). Thus, genes found to be sex-specific in wild-type mice generally retained sex-specificity in the STAT5a-KO mice. Additionally, no discernable overall correlation was found between sex-specificity and the loss of STAT5a in either sex. For 930 of the 3905 genes of interest (24%), the threshold criteria for significant change in expression was met when comparing wild-type and STAT5a-KO mice of the same sex (Table 1). For 579 of these 930 genes (62%), the loss of STAT5a affected gene expression in female liver only, whereas for 289 of the 930 genes (31%), the loss of STAT5a affected gene expression in males only (Table 1). In male mouse liver, STAT5a deficiency had no effect on 93–95% of the sex-specific genes. STAT5a deficiency also had no effect on 93% of male-specific genes in female liver (Table 2). However, of 1437 female-predominant genes, 219 (15%) were decreased in expression in the absence of STAT5a in female liver (Table 2). The response of these female-predominant genes to the loss of STAT5a is discussed further, below.
Table 1. Changes in gene expression in STAT5a-KO mice compared to wild-type mice of the same sex.
930 liver-expressed genes meeting the criteria described in Materials and Methods in either males or females of the STAT5a-knockout strain are listed according to the direction of expression change in knockout (KO) as compared to wild-type (WT) mouse liver and whether the change in expression occurs in males, females or both sexes. 62% of all genes showing a change of expression in any of the STAT5a-knockout mice as compared to wild-type showed a change in expression in females only. 15 genes showed differential expression in both sexes but regulation of these genes by STAT5a was in opposite directions depending upon the sex. Their inclusion in the 930 gene is indicated by an asterisk, but they are not represented in any of the columns.
| Gene expression changes in STAT5a-KO males only | Gene expression changes in STAT5a-KO females only | Gene expression changes in STAT5a-KO mice of both sexes | ||||||
|---|---|---|---|---|---|---|---|---|
| Comparison | Gene Count | Percent (%) | Gene Count | Percent (%) | Gene Count | Percent (%) | Total | Percent (%) |
| WT < KO | 116 | 12 | 231 | 25 | 22 | 2 | 369 | 40 |
| WT > KO | 173 | 19 | 348 | 37 | 25 | 3 | 546 | 59 |
| Total | 289 | 31 | 579 | 62 | 47 | 5 | 930* | |
Table 2. Impact of STAT5a deficiency on male-specific and female-specific gene expression.
2482 liver-expressed genes meeting the criteria for sex-specificity in wild-type mice are listed according to the sex-specificity of their expression in STAT5a-KO mice. Most sex-specific genes do not respond to the loss of STAT5a in either sex, but male genes that do respond tend to be induced in the absence of STAT5a in female liver and repressed in the absence of STAT5a in male liver. In contrast, female genes that respond to the loss of STAT5a tend to be repressed in the absence of STAT5a in female liver. 15% of the female-specific genes show the latter response.
| Response to KO in MalesResponse to KO in Females
|
|||||
|---|---|---|---|---|---|
| Gene Count | Percent (%) | Gene Count | Percent (%) | ||
| Male-specific genes | Increase | 14 | 1 | 56 | 5 |
| Decrease | 61 | 6 | 16 | 2 | |
| No change | 970 | 93 | 973 | 93 | |
| Total | 1045 | 1045 | |||
|
| |||||
| Female-specific genes | Increase | 50 | 3 | 22 | 2 |
| Decrease | 23 | 2 | 219 | 15 | |
| No change | 1364 | 95 | 1196 | 83 | |
| Total | 1437 | 1437 | |||
Clustering by significance and differential expression
The 3905 genes that met the threshold criteria for at least one of the four sex-genotype expression data sets were clustered into subgroups using a “flagging” system (5), whereby each gene is assigned to a specific category based upon its expression ratio in each of the four sex-genotype combinations investigated (see Materials and Methods). 3820 of the 3905 genes were thus classified into the 28 groups of co-expressed genes, shown in Table 3. Table 3 also presents the distribution of gene counts in each of the corresponding categories determined earlier based on sex-specificity and the loss of STAT5b (5). Groups comprised of <15 genes in both the present study and the previous study of STAT5b-KO mice are collected into a single category named “Other”. Six of the seven largest gene groups identified in the present study, comprising 2975 of the 3905 genes (76%) (groups 4A, 4B, 5A, 5B, 7A and 7B) did not show a significant change in expression in either male or female STAT5a-KO mice (i.e., no regulation by STAT5a in either males or females; Table 3, last 2 columns). The remaining 930 genes (24%) met the threshold criteria for a response to STAT5a deficiency in either males or females, as noted above. These genes include group 13B, the sixth largest gene group, which is comprised of 164 female-predominant genes that were down-regulated with STAT5a deficiency in female but not male liver, leading to the loss of sex-specificity in the STAT5a-KO strain (Table 4). A corresponding group of 26 male-predominant genes (group 13A) was specifically up-regulated in STAT5a-deficient female liver, leading to a loss of sex-specificity.
Table 4. Sex-specific liver genes belonging to groups 13A and 13B, which are up- or down-regulated in STAT5a-deficient females but not males and whose sex-specificity is lost in the STAT5a-knockout strain.
Top, shown are the 26 male-specific genes representing group 13A. Bottom, shown are the top 26 female-specific genes representing group 13B. MMT accession numbers are identification numbers assigned to probes that could not be linked to a specific GenBank accession number. Expression ratios shown are average values based on 3 independent microarrays, each carried out in duplicate (dye swap). Individual gene data for all other members of group 13B, and for all other groups listed in Table 3, are presented in Table S2. Values shown in bold are significant by P-value.
| Expression Ratio (average) | ||||||
|---|---|---|---|---|---|---|
| RefSeq/GenBank ID | Common Name(s) | Description | M-WT: F-WT | M-WT: M-KO | F-KO: F-WT | F-KO: M-KO |
| Group 13A. Male predominant and increased in STAT5a-deficient females (total of 26 genes) | ||||||
| NM_146245 | Lrrc21 | leucine rich repeat containing 21 | 3.34 | 0.60 | 4.76 | 0.79 |
| NM_027907 | Agxt2l1 | alanine-glyoxylate aminotransferase 2-like 1 | 3.55 | 0.81 | 3.12 | 0.70 |
| NM_198967 | Tmtc1 | Transmembrane and tetratricopeptide repeat containing 1 | 2.73 | 0.93 | 2.72 | 0.91 |
| NM_025308 | 1810007E14Rik | hypothetical protein LOC53906 | 2.85 | 1.23 | 2.66 | 0.78 |
| MMT00033789 | MMT00033789 | Similar to serine (or cysteine) proteinase inhibitor, clade A, | 2.34 | 1.09 | 2.23 | 1.02 |
| NM_010406 | Hc | hemolytic complement | 3.44 | 2.02 | 2.11 | 1.45 |
| NM_020581 | Angptl4 | angiopoietin-like 4 | 2.18 | 0.82 | 2.06 | 0.74 |
| MMT00033197 | MMT00033197 | Similar to serine (or cysteine) proteinase inhibitor, clade A, | 2.15 | 1.09 | 1.99 | 1.00 |
| NM_008458 | Serpina3c | serine (or cysteine) proteinase inhibitor, clade A, member 3C | 2.00 | 0.86 | 1.96 | 0.79 |
| NM_019748 | Sae1 | SUMO1 activating enzyme subunit 1 | 2.12 | 1.06 | 1.76 | 0.88 |
| NM_145743 | Lace1 | lactation elevated 1 | 1.96 | 0.97 | 1.74 | 0.87 |
| NM_010012 | Cyp8b1 | cytochrome P450, family 8, subfamily b, polypeptide 1 | 2.47 | 1.08 | 1.74 | 0.70 |
| NM_025576 | Ptpmt1 | Protein tyrosine phosphatase, mitochondrial 1 | 1.83 | 1.30 | 1.60 | 1.14 |
| NM_145940 | Wipi1 | WD repeat domain, phosphoinositide interacting 1 | 1.70 | 0.83 | 1.59 | 0.78 |
| NM_172142 | IkappaBNS | NF-kappa B inhibitor | 1.66 | 1.05 | 1.51 | 0.96 |
| NM_134112 | Kctd1 | Potassium channel tetramerisation domain containing 1 | 1.75 | 1.05 | 1.44 | 0.80 |
| NM_019823 | Cyp2d22 | cytochrome P450, family 2, subfamily d, polypeptide 22 | 1.66 | 0.92 | 1.43 | 0.76 |
| NM_133902 | Sds1 | Serine dehydratase-like | 1.53 | 0.94 | 1.40 | 0.83 |
| NM_019911 | Tdo2 | tryptophan 2,3-dioxygenase | 2.08 | 1.05 | 1.39 | 0.82 |
| NM_008391 | Irf2 | interferon regulatory factor 2 | 1.56 | 0.99 | 1.36 | 0.94 |
| NM_001040700 | BC051019 | RefSeq Gene BC051019 | 1.74 | 1.07 | 1.34 | 0.82 |
| NM_172479 | Slc38a5 | Solute carrier family 38, member 5 | 1.53 | 1.01 | 1.34 | 0.92 |
| NM_133906 | Zkscan1 | Zinc finger with KRAB and SCAN domains 1 | 1.60 | 1.23 | 1.30 | 1.01 |
| NM_145365 | Creb3l3 | cAMP responsive element binding protein 3-like 3 | 1.53 | 0.99 | 1.30 | 0.83 |
| NM_008933 | Prm2 | protamine 2 | 1.69 | 1.06 | 1.27 | 0.85 |
| NM_009255 | Serpine2 | serine (or cysteine) proteinase inhibitor, clade E, member 2 | 1.51 | 1.08 | 1.27 | 0.85 |
| Group 13B. Female predominant and decreased in STAT5a-deficient females (total of 164 genes) | ||||||
| NM_175270 | Ankrd56 | Ankrd56 ankyrin repeat domain 56 | 0.27 | 0.76 | 0.13 | 0.40 |
| NM_010766 | Marco | macrophage receptor with collagenous structure | 0.27 | 0.36 | 0.19 | 0.32 |
| NM_007836 | Gadd45a | growth arrest and DNA-damage-inducible 45 alpha | 0.25 | 1.07 | 0.19 | 0.89 |
| NM_010654 | Klrd1 | killer cell lectin-like receptor, subfamily D, member 1 | 0.10 | 1.17 | 0.28 | 1.34 |
| NM_008321 | Idb3 | inhibitor of DNA binding 3 | 0.39 | 0.71 | 0.29 | 0.56 |
| NM_019467 | Aif1 | allograft inflammatory factor 1 | 0.38 | 0.78 | 0.32 | 0.68 |
| NM_011087 | Pira1 | paired-Ig-like receptor A1 | 0.31 | 0.85 | 0.32 | 0.98 |
| NM_020008 | Clec7a | C-type lectin, family 7, member a | 0.40 | 0.93 | 0.34 | 0.85 |
| NM_021272 | Fabp7 | fatty acid binding protein 7, brain | 0.39 | 0.98 | 0.36 | 0.90 |
| NM_010496 | Idb2 | inhibitor of DNA binding 2 | 0.44 | 0.90 | 0.36 | 0.75 |
| NM_010724 | Psmb8 | proteosome subunit, beta type 8 (large multifunctional protease | 0.31 | 0.62 | 0.37 | 0.77 |
| NM_010531 | Il18bp | interleukin 18 binding protein | 0.33 | 0.63 | 0.37 | 0.82 |
| NM_008527 | Klrb1c | killer cell lectin-like receptor subfamily B member 1C | 0.49 | 0.94 | 0.37 | 0.99 |
| BC013712 | BC013712 | basement membrane-induced protein | 0.38 | 0.78 | 0.37 | 0.77 |
| NM_026268 | Dusp6 | dual specificity phosphatase 6 | 0.29 | 1.58 | 0.38 | 2.05 |
| NM_019494 | Cxcl11 | chemokine (C-X-C motif) ligand 11 | 0.45 | 0.84 | 0.39 | 0.90 |
| NM_029620 | Pcolce2 | procollagen C-endopeptidase enhancer 2 | 0.35 | 0.73 | 0.40 | 0.82 |
| NM_010407 | Hck | hemopoietic cell kinase | 0.47 | 0.82 | 0.40 | 0.72 |
| NM_010370 | Gzma | granzyme A | 0.49 | 1.20 | 0.41 | 1.15 |
| NM_178933 | Ostb | organic solute transporter beta | 0.57 | 1.15 | 0.41 | 0.80 |
| NM_030060 | 9130211I03Rik | Jun dimerization protein p21SNFT | 0.35 | 0.83 | 0.41 | 1.10 |
| AF065902 | Itgal | integrin alpha L | 0.37 | 0.99 | 0.41 | 1.21 |
| NM_178759 | Timd4 | T-cell immunoglobulin and mucin domain containing 4 | 0.22 | 0.65 | 0.41 | 1.23 |
| NM_144559 | Fcrl3 | Fc receptor-like 3 | 0.41 | 0.91 | 0.42 | 1.04 |
| NM_172659 | Slc2a6 | solute carrier family 2 (facilitated glucose transporter), member | 0.51 | 0.84 | 0.42 | 0.73 |
| BI407225 | Hk3 | hexokinase 3 | 0.44 | 0.83 | 0.42 | 0.81 |
Female-specific genes that respond to STAT5a deficiency in females
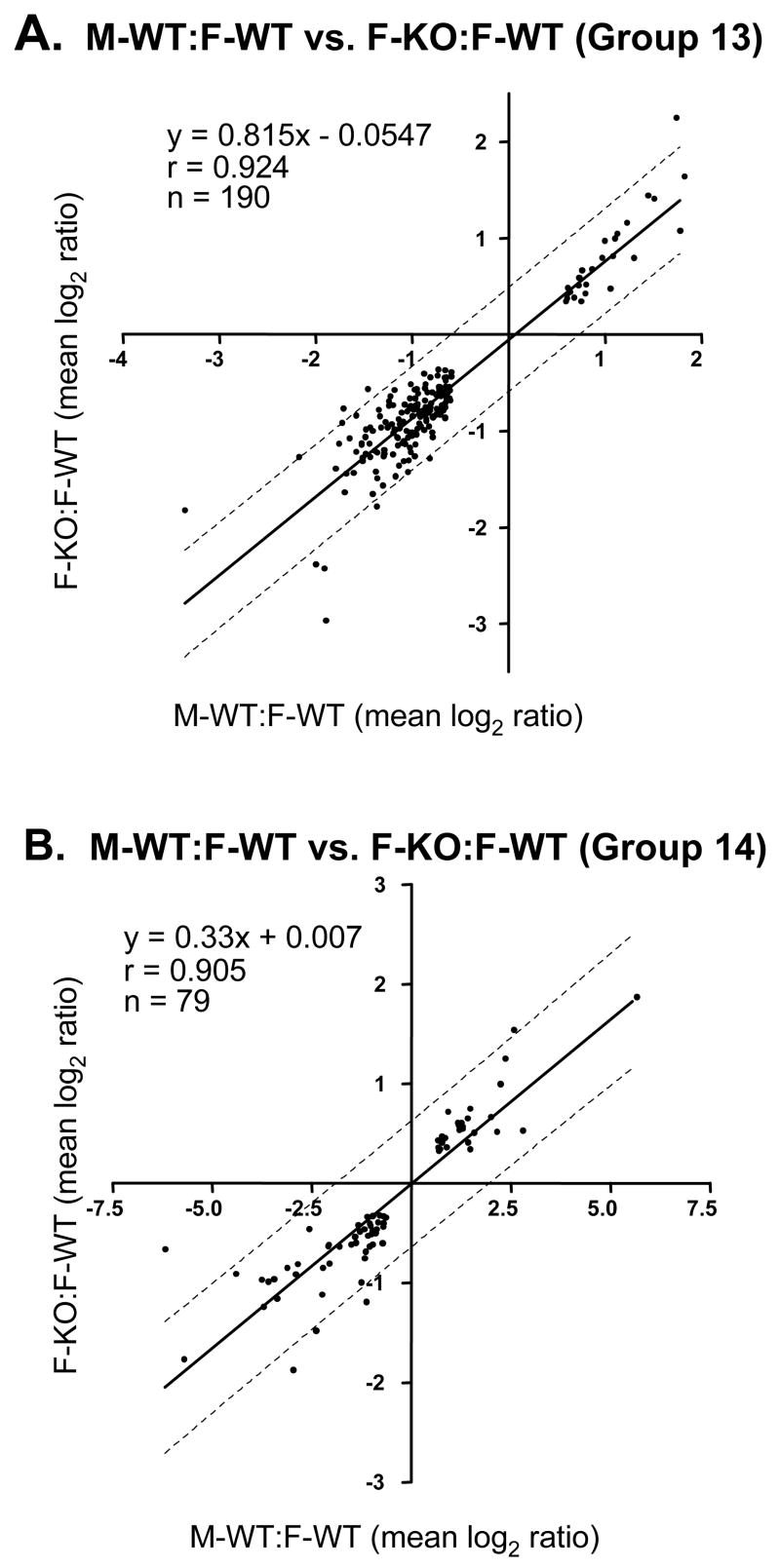
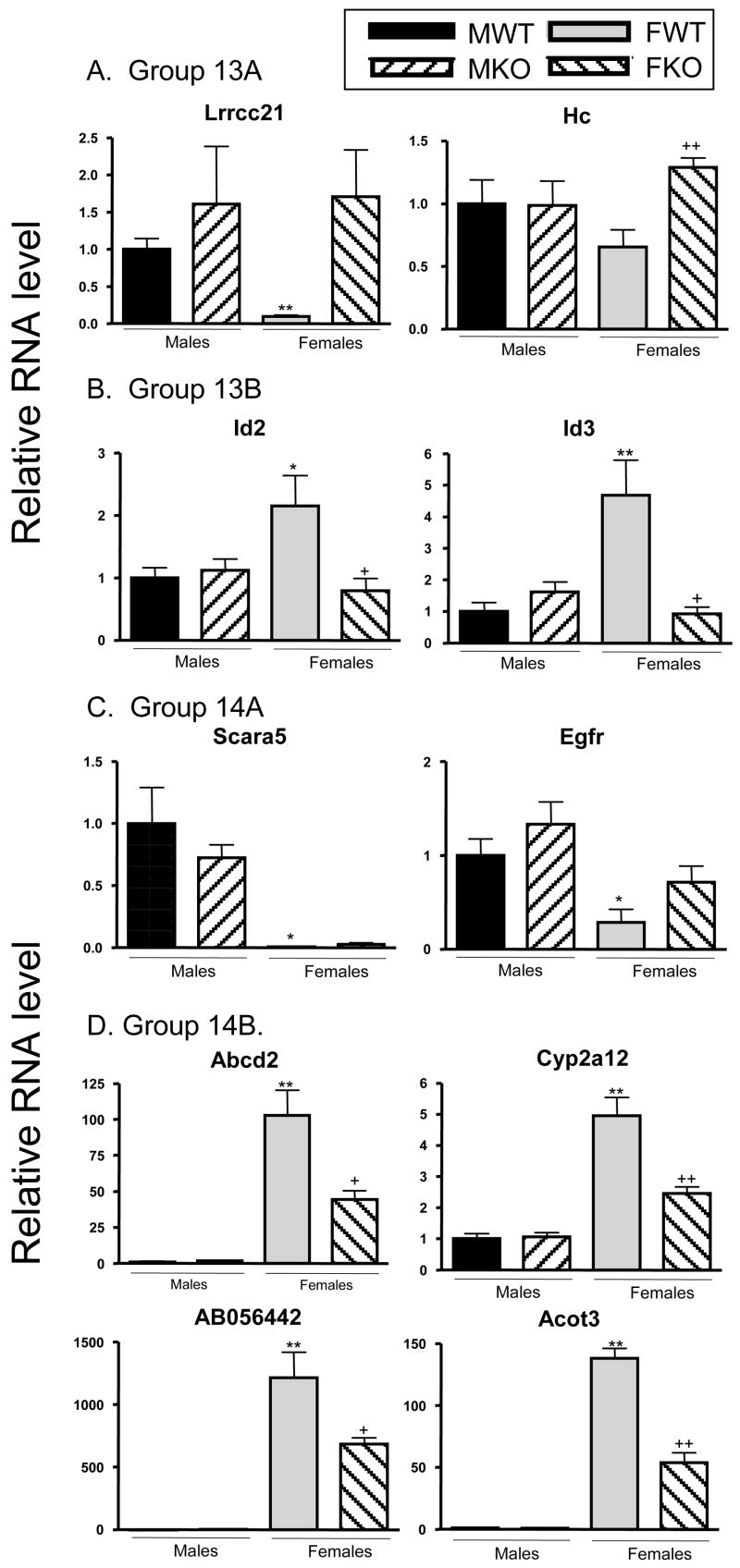
The loss of STAT5a in female liver decreased the sex specificity of 275 genes, with 219 female-predominant genes decreased and 56 male-predominant genes increased (Table 2, column 3). Of these 275 genes, 190 (69%) belong to groups 13A and 13B (Table 3), indicating a loss of sex-specificity in the knockout mice. 79 of the 275 genes (29%) comprise groups 14A and 14B, whose genes display the same pattern of response to STAT5a-deficiency female liver as groups 13A and 13B, albeit with partial retention of sex-specificity in the STAT5a-KO strain (Table 4 vs. Table 5). The quantitative relationship between M-WT:F-WT ratios and F-KO:F-WT ratios for the 190 genes in groups 13A and 13B, and for the 79 genes in groups 14A and 14B, are shown in Fig. 2, where mean log2 ratios for each comparison are plotted on a log-log scale. A linear relationship between the magnitude of sex-specificity and the response to STAT5a deficiency in female liver is evident for the genes in groups 13A and 13B (Fig. 2A; slope = 0.815, y-intercept = 0.0547, r = 0.924) indicating that STAT5a plays a significant role in the sex-specificity of these genes. A linear relationship between sex-specificity and response to the loss of STAT5a in female liver was also seen for the genes in groups 14A and 14B (Fig. 2B; slope = 0.33, y-intercept = −0.007, r = 0.905), where the slope of 0.33 highlights the partial loss of sex-specificity in the absence of STAT5a. These expression profiles were confirmed by qPCR analysis for select genes from groups 13A, 13B, 14A and 14B, as shown in Fig. 3 and in Table S3. A majority of the sex-specific genes responsive to the loss of STAT5a in female liver that are presently annotated with Gene Ontology (GO) terms (194 of the 275 genes in this category) are associated with 6 primary molecular function GO categories: binding, catalytic activity, signal transduction, transporter, enzyme regulation and transcription regulation (Table 6). Four sub-categories of molecular function, signal transducer activity, receptor activity, sugar binding and carbohydrate binding, were significantly enriched (p ≤ 0.0004) among the 194 genes and encompass 68 unique genes.
Table 5. Sex-specific liver genes belonging to groups 14A and 14B, which are up- or down-regulated in STAT5a-deficient females but not males and whose sex-specificity is retained in the STAT5a-knockout strain.
Top, shown are the 28 male-specific genes representing group 14A. Bottom, shown are the top 28 female-specific genes representing group 14B. Other details are as in Table 4. Individual gene data for all other members of group 14B are presented in Table S2. Values shown in bold are significant by P-value.
| Expression Ratio (average) | ||||||
|---|---|---|---|---|---|---|
| RefSeq/GenBank ID | Common Name(s) | Description | M-WT: F-WT | M-WT: M-KO | F-KO: F-WT | F-KO: M-KO |
| Group 14A. Male predominant in WT and KO and increased in STAT5a-deficient females (total of 28 genes) | ||||||
| NM_028903 | Scara5 | Scavenger receptor class A, member 5 (putative) | 50.66 | 0.70 | 3.67 | 0.03 |
| AK050313 | C8b | complement component 8, beta subunit | 6.97 | 0.96 | 1.45 | 0.20 |
| NM_183278 | 2200001I15Rik | RIKEN cDNA 2200001I15 gene | 5.96 | 0.66 | 2.92 | 0.31 |
| NM_007912 | Egfr | epidermal growth factor receptor | 5.15 | 1.14 | 2.39 | 0.53 |
| NM_177101 | 4833442J19Rik | RIKEN cDNA 4833442J19 gene | 4.73 | 0.88 | 2.00 | 0.36 |
| NM_146148 | C8a | complement component 8, alpha polypeptide | 4.44 | ND | 1.44 | 0.24 |
| BC021325 | Serpina1c | Serine (or cysteine) peptidase inhibitor, clade A, member | 4.01 | 0.85 | 1.59 | 0.32 |
| NM_021793 | Tmem8 | transmembrane protein 8 (five membrane-spanning domains) | 3.01 | 1.19 | 1.42 | 0.61 |
| NM_025989 | Gp2 | glycoprotein 2 (zymogen granule membrane) | 2.79 | 1.18 | 1.27 | 0.54 |
| NM_015744 | Enpp2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 | 2.79 | 0.79 | 1.68 | 0.49 |
| BC048380 | Es31 | esterase 31 | 2.68 | 1.07 | 1.33 | 0.51 |
| NM_008645 | Mug1 | murinoglobulin 1 | 2.66 | 0.81 | 1.57 | 0.49 |
| M75720 | Serpina1a | serine (or cysteine) proteinase inhibitor, clade A, member | 2.44 | 0.89 | 1.46 | 0.45 |
| BF147646 | Aak1 | AP2 associated kinase 1 | 2.44 | 0.86 | 1.49 | 0.50 |
| NM_146188 | Kctd15 | potassium channel tetramerisation domain containing 15 | 2.40 | 0.86 | 1.52 | 0.54 |
| NM_013601 | Msx2 | homeo box, msh-like 2 | 2.30 | 0.88 | 1.45 | 0.53 |
| NM_009246 | Serpina1d | serine (or cysteine) proteinase inhibitor, clade A, member | 2.28 | 0.88 | 1.50 | 0.54 |
| NM_009253 | Serpina3m | serine (or cysteine) proteinase inhibitor, clade A, member | 2.25 | 0.86 | 1.53 | 0.60 |
| NM_009695 | Apoc2 | apolipoprotein C-II | 1.90 | 0.79 | 1.65 | 0.64 |
| NM_001039376 | Pde4dip | Phosphodiesterase 4D interacting protein (myomegalin) | 1.85 | 0.97 | 1.29 | 0.70 |
| NM_025680 | Ctnnbl1 | catenin, beta like 1 | 1.81 | 0.98 | 1.37 | 0.74 |
| NM_147217 | Gprc5c | G protein-coupled receptor, family C, group 5, member C | 1.71 | 0.88 | 1.39 | 0.68 |
| NM_010638 | Klf9 | Kruppel-like factor 9 | 1.70 | 0.88 | 1.33 | 0.64 |
| NM_146751 | Olfr648 | olfactory receptor 648 | 1.65 | 0.87 | 1.27 | 0.68 |
| NM_172404 | Ccbl1 | cysteine conjugate-beta lyase 1 | 1.63 | 1.01 | 1.27 | 0.80 |
| NM_172873 | Cdcp2 | CUB domain containing protein 2 | 1.62 | 0.93 | 1.26 | 0.72 |
| NM_011453 | Serpinb9c | Serine (or cysteine) peptidase inhibitor, clade B, member | 1.60 | 0.93 | 1.29 | 0.70 |
| NM_007540 | Bdnf | brain derived neurotrophic factor | 1.60 | 0.89 | 1.35 | 0.63 |
| Group 14B. Female predominant in WT and KO and decreased in STAT5a-deficient females (total of 51 genes) | ||||||
| NM_146232 | BC014805 | hypothetical protein LOC236149 | 0.01 | 0.85 | 0.63 | 33.83 |
| NM_001081325 | EG629219 | Predicted gene, EG629219, sulfotransferase related | 0.02 | 1.02 | 0.29 | 31.19 |
| NM_009286 | C730007P19 | sulfotransferase family 2A, DHEA-preferring, member 2 | 0.05 | 0.91 | 0.53 | 13.85 |
| NM_011994 | Abcd2 | ATP-binding cassette, sub-family D (ALD), member 2 | 0.07 | 0.85 | 0.51 | 6.60 |
| AB056442 | OAT6RP | organic ion transporter 6-related protein | 0.08 | 1.07 | 0.42 | 7.65 |
| NM_007820 | Cyp3a16 | cytochrome P450, family 3, subfamily a, polypeptide 16 | 0.08 | 1.32 | 0.51 | 7.53 |
| MMT00046397 | LOC434121 | sulfotransferase (mSTa1)-related | 0.09 | 1.01 | 0.51 | 5.55 |
| AK149509 | EG233005 | Cytochrome P450, family 2, subfamily a-related | 0.10 | 1.05 | 0.45 | 5.73 |
| NM_134246 | Acot3 | Acyl-CoA thioesterase 3 | 0.12 | 1.10 | 0.56 | 5.92 |
| NM_054088 | Pnp1a3 | Patatin-like phospholipase domain containing 3 | 0.13 | 1.42 | 0.27 | 3.33 |
| NM_007643 | Cd36 | CD36 antigen | 0.13 | 0.72 | 0.53 | 2.93 |
| NM_018784 | St3gal6 | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 0.14 | 0.87 | 0.57 | 4.22 |
| NM_008706 | Nqo1 | NAD(P)H dehydrogenase, quinone 1 | 0.17 | 0.81 | 0.73 | 3.94 |
| NM_178765 | 5730410E15Rik | Golgi-localized syntaphilin-related protein isoform C | 0.19 | 1.11 | 0.36 | 3.27 |
| NM_008935 | Prom1 | prominin 1 | 0.21 | 1.06 | 0.46 | 2.67 |
| NM_008357 | Il15 | interleukin 15 | 0.22 | 0.83 | 0.56 | 2.28 |
| BB153684 | Slc16a5 | solute carrier family 16 (monocarboxylic acid transporters), | 0.24 | 1.07 | 0.65 | 3.54 |
| NM_019919 | Ltbp1 | latent transforming growth factor beta binding protein 1 | 0.24 | 0.79 | 0.65 | 2.40 |
| NM_013825 | Ly75 | lymphocyte antigen 75 | 0.24 | 0.76 | 0.57 | 1.91 |
| NM_009982 | Ctsc | cathepsin C | 0.29 | 0.80 | 0.64 | 1.82 |
| NM_029653 | Dapk1 | death associated protein kinase 1 | 0.35 | 1.46 | 0.65 | 2.89 |
| NM_025408 | Phca | phytoceramidase, alkaline | 0.38 | 1.24 | 0.69 | 2.38 |
| NM_144846 | 0910001A06Rik | hypothetical protein LOC223601 | 0.38 | 1.12 | 0.66 | 1.94 |
| NM_023737 | Ehhadh | hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 0.40 | 1.18 | 0.75 | 2.25 |
| NM_007987 | Fas | Fas (TNF receptor superfamily member) | 0.41 | 0.94 | 0.72 | 1.61 |
| NM_008371 | Il7 | interleukin 7 | 0.42 | 1.04 | 0.50 | 1.57 |
| NM_026082 | Dock7 | dedicator of cytokinesis 7 | 0.44 | 0.97 | 0.73 | 1.64 |
| NM_126166 | Tlr3 | toll-like receptor 3 | 0.44 | 1.03 | 0.59 | 1.39 |
Fig. 2. Scatterplots for quantitative correlation of M-WT:F-WT and F-KO:F-WT ratios for genes in groups 13 and 14.
A, Shown are log2 ratios for M-WT:F-WT (x-axis) and the corresponding F-KO:F-WT data (y-axis) plotted on a log-log scale for all genes in groups 13A and 13B (Table 3; n=190 genes). The two sets of ratios for the two experimental comparisons are highly correlated (r = 0.924) and the best-fit line (y = 0.815x − 0.0547), shown in solid, has a slope close to 1 and an intercept near 0. The 95% prediction boundaries are shown as dashed lines. B, Shown are log2 ratios for M-WT:F-WT (x-axis) and the corresponding F-KO:F-WT data (y-axis) plotted on a log-log scale for all genes in groups 14A and 14B (Table 3; n=79 genes). The two sets of ratios for the two experimental comparisons are highly correlated (r = 0.905) and the best-fit line (y = 0.33x + 0.007), shown in solid, has an intercept near 0 but a slope near one-third. The 95% prediction boundaries are shown as dashed lines. In both panels, genes found in the upper right quadrant (I) are male-predominant and were increased in expression in female liver in the absence of STAT5a, while genes found in the lower left quadrant (III) are female-predominant and were decreased in expression in STAT5a-deficient female liver.
Fig. 3. qPCR analysis of select genes in groups 13A, 13B, 14A and 14B.
RNA samples prepared from individual male and female livers, wild-type (WT) and STAT5a-knockout (KO), as indicated, were assayed for the indicated RNAs from each group as described under Materials and Methods. Significant differences are indicated as follows. For F-WT vs. M-WT: *, p<0.05 and **, p<0.01; for M-KO vs M-WT and F-KO vs F-WT; +, p<0.05 and ++, p<0.01. Results confirm the general trends associated with each group (c.f., Tables 4 and 5).
Table 6. Distribution of molecular function-related GO categories and GO enrichment for 194 sex-specific genes that show a dependence on STAT5a in female mouse liver.
A, Genes showing sex-specific expression (M-WT:F-WT >1.5 or <0.66 and p < 0.05) and a response to the loss of STAT5a in female liver (F-KO:F-WT > 1.25 or <0.8 and p<0.05) were grouped within GeneSpring GX on the basis of the molecular function-related GO categories assigned to the genes. 194 genes met the criteria for inclusion and were annotated with the GO categories shown. Genes were also assigned all parent categories within the ontology. Category numbers marked with an asterisk were enriched and are presented in more detail in part B. B, Enriched GO terms were selected for over-representation of genes with P < 0.0001. The total number of genes and the percentage of total genes within each GO category relates to all genes represented on the microarray with the indicated GO annotations. The select gene count and the percentage of selected genes in each category are those genes within the 194 genes described above within the indicated GO category. P values were assigned by GeneSpring GX for each GO category based on whether the number of selected genes within each category was greater than expected by randomly sampling 194 genes from the total set of genes represented on the microarray given the subset of the total number of genes with that GO annotation. Child categories are noted below their parent categories within the GO hierarchy.
| A. GO Category Distribution | GO Category Number | Gene Count | Percentage | ||
|---|---|---|---|---|---|
| Binding | 5488* | 101 | 35.4 | ||
| Catalytic activity | 3824 | 65 | 22.8 | ||
| Signal transducer activity | 4871* | 63 | 22.1 | ||
| Transporter activity | 5215 | 16 | 5.6 | ||
| Enzyme regulator activity | 30234 | 16 | 5.6 | ||
| Transcription regulator activity | 30528 | 11 | 3.9 | ||
| Structural molecule activity | 5198 | 2 | 0.7 | ||
| Antioxidant activity | 16209 | 1 | 0.4 | ||
| Molecular function unknown | 5554 | 10 | 3.5 | ||
| B. Enriched GO terms | |||||
| GO category | Total genes in category | Percent of total genes in category %) | Select genes in Category | Percent of select genes in category (%) | P value |
|
| |||||
| GO:4871: signal transducer activity | 2550 | 19.48 | 63 | 32.47 | 0.00001 |
| -- GO:4872: receptor activity | 1960 | 14.97 | 51 | 26.29 | 0.00003 |
| GO:5488: Binding | |||||
| -- GO:30246: carbohydrate binding | 232 | 1.77 | 13 | 6.70 | 0.00004 |
| -- GO:5529: sugar binding | 167 | 1.28 | 11 | 5.67 | 0.00004 |
Comparison of STAT5a deficiency to STAT5b deficiency
No correlation between sex-specific gene expression and the effect of STAT5a deficiency was seen, either in males or in females, in contrast to our earlier finding with STAT5b deficient mice (5). Qualitatively, when the regulated genes in the present STAT5a-KO study were sorted by sex-specificity, only the M-KO:F-KO comparison showed similarity to M-WT:F-WT in the false-color heat map (Fig. 1, lane 2 vs. lane 1), whereas an extensive similarity in the heatmap was apparent for the M-WT:F-WT and M-WT:M-KO comparisons in the STAT5b-KO study (lane 8 vs. lane 5). The distribution of genes among the co-expressed gene groups also exhibited marked differences in the requirement of STAT5a and STAT5b for sex-specific gene expression (Table 3). STAT5b was required for expression in male liver of 1205 sex-specific genes, or 75% of the 1603 sex-specific genes identified in the STAT5b study (5) (genes comprising STAT5b-KO study groups 1A, 1B, 2A, 2B, 3A, 3B, 6A and 6B). By contrast, the corresponding eight groups comprised ~ 4% of the 2482 sex-specific genes in the present STAT5a study (Table 3). Six of the seven largest groups of co-expressed genes identified in the present study are sex-specific genes that did not respond to the loss of STAT5a in either males or females (groups 4A, 4B, 5A, 5B, 7A, and 7B). Together, these six groups comprise 76% of the genes of interest in the STAT5a study vs. only 23% of the corresponding set of genes in the STAT5b study. Major differences in the impact of STAT5a vs. STAT5b deficiency are thus apparent.
Comparison of sex-specificity among three mouse strains
M-WT:F-WT expression ratios, obtained in the present analysis of 129J × Black Swiss mouse livers, were compared to the corresponding ratios determined using the same microarray platform for two sets of male and female mouse livers from different genetic backgrounds: 129 × BALB/c mice (genetic background used in the STAT5b-KO mouse study (5)), and ICR mice, an outbred strain. Normalized M-WT:F-WT expression ratios for a total of ten microarrays (3 from 129J × Black Swiss mice, 3 from 129 × BALB/c mice, and 4 from ICR mice) were combined to obtain a single data set, of which 1028 genes met the threshold of >1.5 fold differential expression at p<0.001 (Table S4). 523 of these 1028 genes met the threshold of >1.5-fold differential expression and p<0.05 in all three independent data sets, indicating robust sex-specific expression across strains and experiments (Fig. S1 and Table S5).
Further examination of the three microarray data sets revealed genes that met the criteria for sex-specificity in only one of the data sets (Fig. S1, outer, non-overlapping portions of each circle). ANOVA analysis identified 54 genes showing sex-specificity in 129 × BALB/c mice, but not in the other two strains, at p<0.001 (Table S6). Similarly, 49 genes were identified as sex-specific in ICR mice only, while 551 genes were sex-specific in 129J × Black Swiss mice only. Top candidates for these strain-dependent, sex-specific genes are presented in Table S6.
Discussion
The present study investigates the role of STAT5a in the sexual dimorphism of mouse liver gene expression, which was previously shown to be extensive and highly dependent on STAT5b, the most abundant liver STAT5 form. Whereas a majority of the sex-specific hepatic genes identified here were unaffected by the loss of STAT5a, expression of a distinct subset of these genes was specifically altered in STAT5a-deficient female mouse liver. These include four female-specific Cyp genes, Cyp2a12, Cyp2d22, Cyp3a16, and Cyp8b1, all of which are dependent on STAT5a for expression in female liver (gene groups 13 and 14; Table 3). Overall, 219 genes, corresponding to 15% of female-predominant genes, were decreased in expression while 5% of male-predominant genes were increased in expression in livers of female but not male mice deficient in STAT5a. The portion of female-predominant genes that required STAT5a for expression increased to 23% (91 genes) when a strain-independent set of 405 female-specific genes was considered. This finding contrasts with the extensive regulation of sex-specific gene expression by STAT5b in male liver, where nearly 90% of all sex-specificity is lost or substantially reduced in the absence of STAT5b (5). Moreover, the distribution of genes among the threshold-based groups between STAT5b and STAT5a were significantly different (Table 3). Interestingly, four molecular functions as annotated by GO were over-represented in the set of sex-specific genes regulated by STAT5a in female liver: signal transduction, receptors, carbohydrate binding, and sugar binding (p < 0.0001).
Earlier studies suggested a role for STAT5a in the expression of a female-specific Cyp2b family member, based on the loss of a female-specific Cyp2b protein in STAT5a-deficient female mouse liver (20). In that case, however, the Cyp2b protein was also decreased in STAT5b-deficient females, a pattern of regulation distinct from that of the major group of STAT5a-dependent female genes described here. The role of STAT5a as a feminizing factor in female mouse liver reported here (positive regulation of female genes and negative regulation of male genes) is analogous to that of STAT5b in male liver, where STAT5b serves as a masculinizing factor (positive regulation of male genes and negative regulation of female genes). STAT5a but not STAT5b also plays the major role in another female-specific function – mammary gland differentiation and lactogenesis in response to prolactin stimulation (15).
A majority of the differentially expressed genes identified in the present STAT5a data set (2482 of the 3905 genes; 64%) exhibited sex-specific expression in wild-type mouse liver. This proportion is similar to the 1603 of 2231 differentially expressed genes (72%) identified in our earlier study of STAT5b-KO mice (5). However, only 659 genes overlap between these two groups (i.e., the 2482 sex-dependent genes from the STAT5a dataset, based on 129J x Black Swiss mice, and 1603 sex-dependent genes from the STAT5b dataset, based on 129 x BALB/c mice). Our analysis of sex-specificity in a third mouse strain, ICR, using the same microarray platform yielded 1810 sex-specific genes, 1135 of which were sex-specific in at least one of the other two strains (Fig. S1). Analysis of the sex-specificity data from all three strains (10 microarrays) as a single data set identified 1028 genes showing sex-dependent expression at p<0.001 across all three studies (Table S4). These include liver-expressed Y-linked genes such as Ddx3y, Eif2s3y, and Jarid1d, as well as 26 Cyp genes and 10 Sult (sulfotransferase) genes. A subset comprised of 523 genes met the threshold for sex-specificity in all three strain backgrounds, and thus correspond to a robust core of sex-specific hepatic genes. Forty-seven genes annotated as transcriptional regulators by GO were included among the robust sex-specific genes, including cut-like 2 (Cutl2), SRY-box containing gene 15 (Sox15), and myogenic factors 5 and 6 (Myf5, Myf6), a subset of which may mediate the sex-specific actions of GH-activated STAT5a or STAT5b, as discussed elsewhere (29).
We also identified genes whose sex-specificity was apparent in livers from only one of the three genetic backgrounds investigated. Some of these genes may be subject to strain-dependent genetic regulation. Genes exhibiting significant discrepancies in expression across the three strain backgrounds are listed in Table S6. Only a subset of the genes found to be sex-specific in a single strain background is likely to exhibit true strain-dependent sex-specificity. Technical factors inherent to microarray analysis, as well as the fact that individual livers, rather than pools of livers, comprised the biological triplicates for one of the microarray studies, may contribute to the apparent strain-dependent loss of sex-specificity for some of these genes. Nevertheless, these findings suggest that the strain-dependence of liver sex-specificity may be more extensive than was previously recognized. Previous studies identified Rsl alleles that conferred strain-specific regulation of certain sex-specific genes, notably Slp, Cyp2d9, and several Mup family members (14). In other studies, strain-specific regulation of the female-specific Cyp2a4 was associated with a recessive mutation at the GH-dependent repression (Gdr) locus abolishes repression of Cyp2a4 expression in males (2). Further investigation using larger numbers of arrays representing several pure outbred and inbred mouse strains, including strains containing the Rsl and Gdr mutations, will be necessary to isolate effects due to strain differences from those due to strain-independent individual variability and technical limitations of microarray technology.
It cannot be determined whether the STAT5a dependence of sex-specific gene expression seen here in female liver reflects a direct role of STAT5a, or whether the requirement for STAT5a is indirect. It is also unclear why STAT5b is unable to compensate for the loss of STAT5a, given its much higher expression in liver (20) and the very close similarities in biochemical and gene regulatory activities of these two closely related (>90% identical) STAT5 proteins (7, 11). STAT5b has been proposed to serve as an upstream transcriptional regulator of a larger, downstream transcriptional network that leads to activation of many of the male-predominant genes and repression of the many female-predominant genes whose hepatic expression is dependent on STAT5b (29). Conceivably, STAT5a could play such a role, albeit on a much smaller scale, in female liver. Of note, eleven of the sex-specific genes dependent on STAT5a in female liver were annotated as transcriptional regulators by GO; these include Inhibitor of DNA binding 2 (Id2) and Interferon regulatory factor 2 (Irf2). The apparent STAT5a-dependent repression of male-predominant genes in female liver might not be a direct function of STAT5a. For example, an inhibitory nuclear factor, such as Id2, could serve as a direct target of STAT5a and contribute to STAT5a inhibition of select male-specific genes in female liver. Interestingly, Id2 and STAT5a show cooperative activity in development and maturation of mouse mammary tissue, where the loss of Id2 decreased active, tyrosine phosphorylated STAT5a levels by 40% (18). A similar pathway, involving STAT5a and other transcriptional regulators, could be responsible for the limited number of genes that exhibit STAT5a dependence in female liver. Further studies will be required to elucidate the role of STAT5a in female gene expression in liver and other tissues, including mammary gland, where STAT5a is the major STAT5 form.
Supplementary Material
Acknowledgments
Supported in part by NIH grant DK33765 (to D.J.W.). K.H.C. and G.D.M. received Training Core support from the Superfund Basic Research Center at Boston University, NIH Grant 5 P42 ES07381, and V.W. received fellowship support from the Belgian American Educational Foundation.
Abbreviations
- STAT
signal transducer and activator of transcription
- GH
growth hormone
- CYP
cytochrome P450
- WT
wild-type
- STAT5a-KO
STAT5a knockout
- STAT5b-KO
Stat5b knockout
- GO
Gene Ontology
Footnotes
Supported in part by NIH grant DK33765 (to DJW)
References
- 1.Ahluwalia A, Clodfelter KH, Waxman DJ. Sexual dimorphism of rat liver gene expression: regulatory role of growth hormone revealed by deoxyribonucleic Acid microarray analysis. Mol Endocrinol. 2004;18:747–760. doi: 10.1210/me.2003-0138. [DOI] [PubMed] [Google Scholar]
- 2.Aida K, Negishi M. A trans-acting locus regulates transcriptional repression of the female-specific steroid 15 alpha-hydroxylase gene in male mice. Journal of molecular endocrinology. 1993;11:213–222. doi: 10.1677/jme.0.0110213. [DOI] [PubMed] [Google Scholar]
- 3.Choi HK, Waxman DJ. Growth hormone, but not prolactin, maintains, low-level activation of STAT5a and STAT5b in female rat liver. Endocrinology. 1999;140:5126–5135. doi: 10.1210/endo.140.11.7106. [DOI] [PubMed] [Google Scholar]
- 4.Choi HK, Waxman DJ. Plasma growth hormone pulse activation of hepatic JAK-STAT5 signaling: developmental regulation and role in male-specific liver gene expression. Endocrinology. 2000;141:3245–3255. doi: 10.1210/endo.141.9.7638. [DOI] [PubMed] [Google Scholar]
- 5.Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol. 2006;20:1333–1351. doi: 10.1210/me.2005-0489. [DOI] [PubMed] [Google Scholar]
- 6.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 7.Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–157. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 8.Herrington J, Carter-Su C. Signaling pathways activated by the growth hormone receptor. Trends Endocrinol Metab. 2001;12:252–257. doi: 10.1016/s1043-2760(01)00423-4. [DOI] [PubMed] [Google Scholar]
- 9.Holloway MG, Laz EV, Waxman DJ. Codependence of growth hormone-responsive, sexually dimorphic hepatic gene expression on signal transducer and activator of transcription 5b and hepatic nuclear factor 4alpha. Mol Endocrinol. 2006;20:647–660. doi: 10.1210/me.2005-0328. [DOI] [PubMed] [Google Scholar]
- 10.Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 11.Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. 2001;13:211–217. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 12.Jansson JO, Eden S, Isaksson O. Sexual dimorphism in the control of growth hormone secretion. Endocr Rev. 1985;6:128–150. doi: 10.1210/edrv-6-2-128. [DOI] [PubMed] [Google Scholar]
- 13.Krebs CJ, Larkins LK, Khan SM, Robins DM. Expansion and diversification of KRAB zinc-finger genes within a cluster including Regulator of sex-limitation 1 and 2. Genomics. 2005;85:752–761. doi: 10.1016/j.ygeno.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Krebs CJ, Larkins LK, Price R, Tullis KM, Miller RD, Robins DM. Regulator of sex-limitation (Rsl) encodes a pair of KRAB zinc-finger genes that control sexually dimorphic liver gene expression. Genes Dev. 2003;17:2664–2674. doi: 10.1101/gad.1135703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 16.MacLeod JN, Pampori NA, Shapiro BH. Sex differences in the ultradian pattern of plasma growth hormone concentrations in mice. J Endocrinol. 1991;131:395–399. doi: 10.1677/joe.0.1310395. [DOI] [PubMed] [Google Scholar]
- 17.Mode A, Gustafsson JA. Sex and the liver - a journey through five decades. Drug Metab Rev. 2006;38:197–207. doi: 10.1080/03602530600570057. [DOI] [PubMed] [Google Scholar]
- 18.Mori S, Nishikawa SI, Yokota Y. Lactation defect in mice lacking the helix-loop-helix inhibitor Id2. Embo J. 2000;19:5772–5781. doi: 10.1093/emboj/19.21.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noshiro M, Serabjit-Singh CJ, Bend JR, Negishi M. Female-predominant expression of testosterone 16 alpha-hydroxylase (“I”-P-450(16)alpha) and its repression in strain 129/J. Arch Biochem Biophys. 1986;244:857–864. doi: 10.1016/0003-9861(86)90655-7. [DOI] [PubMed] [Google Scholar]
- 20.Park SH, Liu X, Hennighausen L, Davey HW, Waxman DJ. Distinctive roles of STAT5a and STAT5b in sexual dimorphism of hepatic P450 gene expression. Impact of STAT5a gene disruption. J Biol Chem. 1999;274:7421–7430. doi: 10.1074/jbc.274.11.7421. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz J, Huo JS, Piwien-Pilipuk G. Growth hormone regulated gene expression. Minerva Endocrinol. 2002;27:231–241. [PubMed] [Google Scholar]
- 22.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 23.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 24.Tullis KM, Krebs CJ, Leung JY, Robins DM. The regulator of sex-limitation gene, rsl, enforces male-specific liver gene expression by negative regulation. Endocrinology. 2003;144:1854–1860. doi: 10.1210/en.2002-0190. [DOI] [PubMed] [Google Scholar]
- 25.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veldhuis JD, Anderson SM, Shah N, Bray M, Vick T, Gentili A, Mulligan T, Johnson ML, Weltman A, Evans WS, Iranmanesh A. Neurophysiological regulation and target-tissue impact of the pulsatile mode of growth hormone secretion in the human. Growth Horm IGF Res. 2001;11 (Suppl A):S25–37. doi: 10.1016/s1096-6374(01)80005-8. [DOI] [PubMed] [Google Scholar]
- 27.Waxman DJ. Growth hormone pulse-activated STAT5 signalling: a unique regulatory mechanism governing sexual dimorphism of liver gene expression. Novartis Found Symp. 2000;227:61–74. doi: 10.1002/0470870796.ch5. discussion 75–81. [DOI] [PubMed] [Google Scholar]
- 28.Waxman DJ, Celenza JL. Sexual dimorphism of hepatic gene expression: novel biological role of KRAB zinc finger repressors revealed. Genes Dev. 2003;17:2607–2613. doi: 10.1101/gad.1154603. [DOI] [PubMed] [Google Scholar]
- 29.Waxman DJ, O’Connor C. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol. 2006;20:2613–2629. doi: 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- 30.Waxman DJ, Ram PA, Park SH, Choi HK. Intermittent plasma growth hormone triggers tyrosine phosphorylation and nuclear translocation of a liver-expressed, Stat 5-related DNA binding protein. Proposed role as an intracellular regulator of male-specific liver gene transcription. J Biol Chem. 1995;270:13262–13270. doi: 10.1074/jbc.270.22.13262. [DOI] [PubMed] [Google Scholar]
- 31.Wiwi CA, Waxman DJ. Role of hepatocyte nuclear factors in growth hormone-regulated, sexually dimorphic expression of liver cytochromes P450. Growth Factors. 2004;22:79–88. doi: 10.1080/08977190410001715172. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.