Fig. 6.
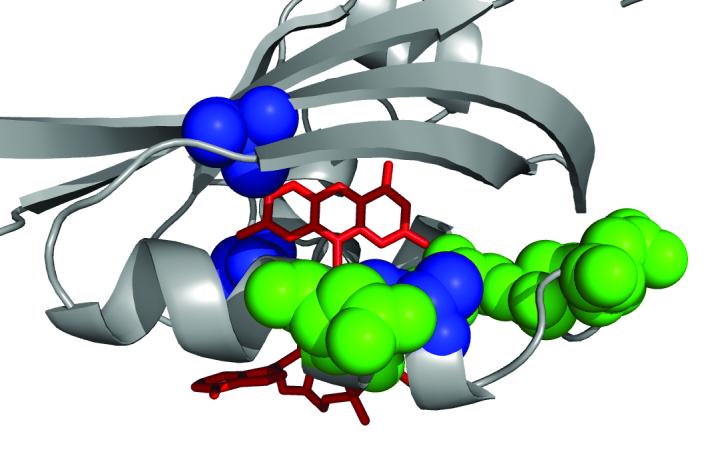
A view of the FAD-binding cavity in Aer PAS showing residues critical for FAD binding (green), and location of general suppressors (blue) of HAMP[AS-2] null mutants. FAD coordinates are modeled on the crystal structure of the NifL protein.

A view of the FAD-binding cavity in Aer PAS showing residues critical for FAD binding (green), and location of general suppressors (blue) of HAMP[AS-2] null mutants. FAD coordinates are modeled on the crystal structure of the NifL protein.