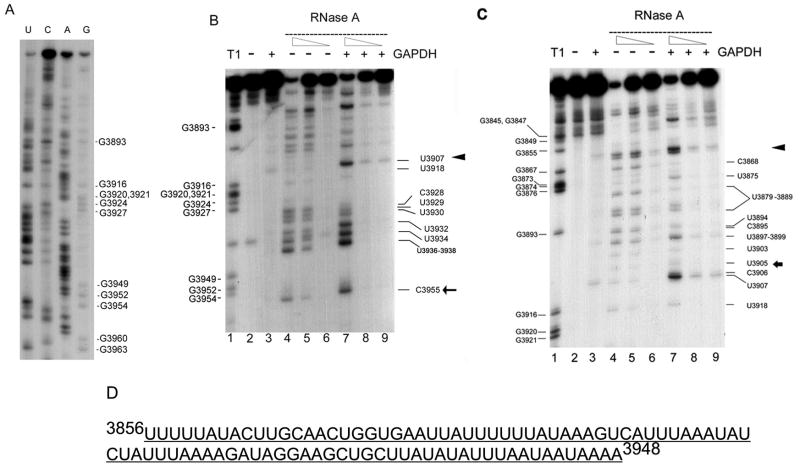
Figure 3. RNA footprinting analysis of the interaction of GAPDH protein with the 3′ untranslated region of CSF-1 RNA.
(A) 3′ end-labeled 3′ UTR CSF-1 RNA (3829-3972nt) sequenced chemically. The pattern derived was used to help interpret the footprinting experiments. (B) 3′ region of GAPDH binding to the 3′UTR CSF-1 RNA. The lane numbers are at the bottom of each panel. Lane 1, labeled CSF-1 RNAs (3828-3972nt) are partially hydrolyzed by RNase T1 (cleavage site 3′ of single-stranded G’s) to provide a reference ladder. CSF-1 RNA (3828-3972 nt) was subjected to a standard binding reaction in the presence (lane 3, 7, 8, 9) (+) or absence (lane 2, 4, 5, 6) (−) of 1.4 μg human GAPDH, followed by cleavage with RNase A (lanes 4–9). Partial cleavage with the decreasing amounts of RNase A (0.1ng/ml (lanes 4, 7), 0.02ng/ml (lanes 5, 8), 0.004ng/ml (lanes 6, 9)) or control reactions without RNase (lanes 2, 3) was performed. Triangle and arrow indicate the approximate boundaries of the 3′ region of the footprint between (3908-3916nt) and 5′ to 3955nt (the 3′ end of the footprint is better defined as 3939-3948nt in the text). (C) 5′ region of GAPDH binding to the 3′UTR CSF-1 RNA. The conditions for each lane are as described in (B). Triangle and arrow indicate the approximate boundaries of the 5′ region of the footprint between (3856-3868nt) and 3905nt. The 5′ region of the footprint can also be visualized in (B), but with less detail. (D) AU-rich footprint for GAPDH binding of 3′UTR CSF-1 RNA.