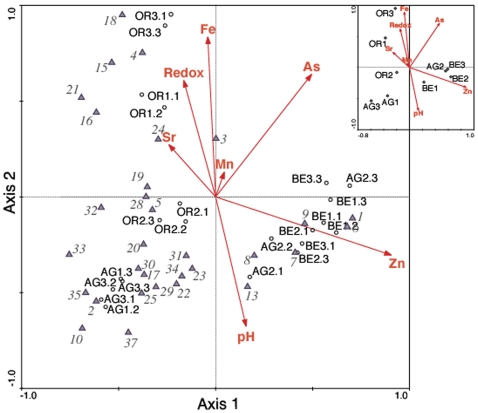
Figure 6. CCA biplot of the SARST-V6 dataset with relevant environmental variables at Río Tinto samples and sites.
Superimposed canonical correspondence analysis (CCA) biplots of RT samples and SARST-V6 OTUs at the 99% similarity cut-off value displaying 68% of the variance of the OTUs with respect to the environmental variables. The inset represents the CCA biplot when pooling samples by site. The canonical eigenvalues for axes 1–4 of the sample analysis are 0.367, 0.272, 0.112, and 0.062 respectively. Environmental variables are indicated by arrows that point in the direction of increasing values of each variable. The coordinates of the arrowheads indicate the degree of correlation with the axes. Samples and sites are represented by black circles. For sample names see Materials and Methods. OTUs with total abundances higher than 10 RSTs are represented by grey triangles. To avoid overcrowding of points only one OTU per strain is plotted. The relative frequency of OTUs in samples can be determined using the biplot rule. To do this, drop a perpendicular from each sample onto a line through the OTU and the origin. Samples projecting on the line in the direction towards the OTU and beyond it are predicted to have a higher relative frequency of that OTU than samples projecting onto the line in the opposite direction. Interpretation of environmental arrows with respect to sites, OTUs and other environmental variables follows the same rule. OTU numbers correspond to: (1, 12, 14, 36) = Acidithiobacillus sp. SS5; (2, 11) = Uncultured bacterial clone MPKCSC9; (3) = Acidithiobacillus sp. SK5; (4) = Leptospirillum ferrooxidans P3a; (5, 26) = L. ferrooxidans Parys; (6) = Acidithiobacillus sp. B9; (7) = L. ferrooxidans Sy; (8) = Thermicanus aegyptius; (9) = Acidiphilium sp. Pk46; (10) = Eubacterium clones TRA5-3 and MeBr10; (13) = Uncultured bacterium BA18; (15) = F. acidiphilium; (16) = Bacterium clone 015C-C11; (17) = Actinomycetales clone TM167; (18) = Leptospirillum sp. strain DSM 2391; (19) = Thermicanus aegyptius; (20) = Bacterium Ellin5017; (21) = Pseudomonas sp. B35; (22) = Nostoc sp. PCC 9231; (23) = Acidiphilium sp. CCP3; (24) = Uncultured bacterium clone RCP2-12; (25) = Uncultured actinobacterium clone BPM2_A01; (27) = Acidithiobacillus sp. SK5; (28) = Acidobacteria clone BPC3_E10; (29) = Uncultured bacterium clone 300A-B12; (30) = Bacterium Ellin5114; (31) = Corynebacterium sp. S18-03; (32) = Uncultured bacterium clone RCP1-34; (33) = Uncultured bacterium clone RH1-L2; (34) = Uncultured bacterium clone RH1-i3; (35) = Uncultured bacterium clone RCP2-16; (37) = Uncultured actinobacterium clone BPM3_G08.