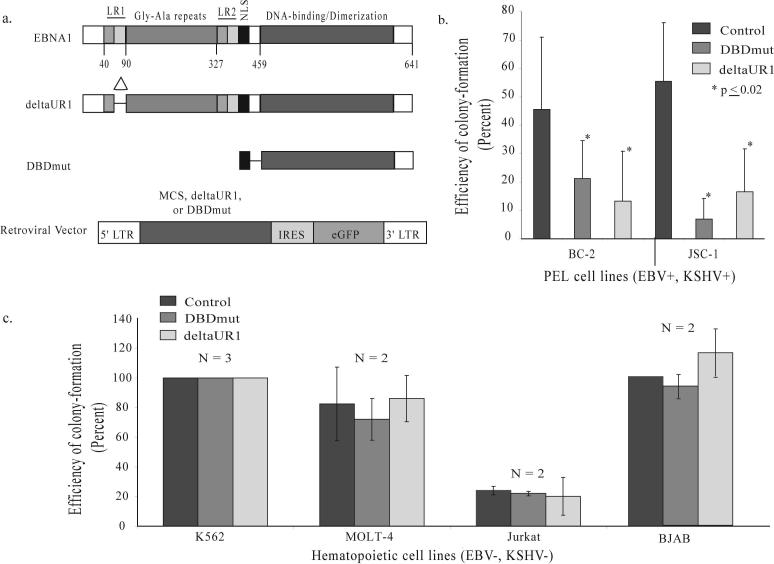
Figure 1. Two derivatives of EBNA1 inhibit EBV+, KSHV+ PEL cells from forming colonies and do not affect EBV−, KSHV− hematopoietic cell lines.
(a) Full-length EBNA1 is 641 amino acids in length, contains two regions (LR1 and LR2) responsible for linking DNA, a nuclear localization sequence (NLS) and a DNA-binding and dimerization domain (15). This carboxy-terminal DNA-binding and dimerization domain is not sufficient to support EBNA1's contributions to DNA replication and acts as a dominant-negative derivative inhibiting all of these functions (9, 15). Numbers indicate the position of amino acids. The deletion within deltaUR1 spans amino acids 65 − 89 (Delta) rendering EBNA1 transcriptionally defective. DBDmut contains only an NLS and the DNA-binding and dimerization domain and is defective in all of EBNA1's known functions. DBDmut, deltaUR1, or a multiple cloning sequence (MCS) as the control was inserted into a retroviral backbone as previously described (9) that co-expresses eGFP. (b) BC-2 and JSC-1 cells were infected with retroviruses expressing DBDmut, deltaUR1, or the control retrovirus containing only the MCS. Infected cells were sorted by FACs 48 hours post-infection, and their efficiency of forming colonies determined 2 weeks post-plating. (c) DBDmut and deltaUR1 do not inhibit EBV(−), KSHV(−) hematopoietic cell lines from forming colonies. Two EBV-negative Burkitt's lymphoma cell lines BJAB and DG75 (analyzed in reference 9); an erythroleukemia cell line (K562); and two T-cell lymphoblastic leukemia lines (MOLT-4 and Jurkat) were tested in assays for colony-formation. These cells were infected with control, DBDmut, or deltaUR1-expressing retroviruses, sorted into 96-well plates and measured for their ability to form colonies 2 weeks post-plating. N, number of independently performed experiments.