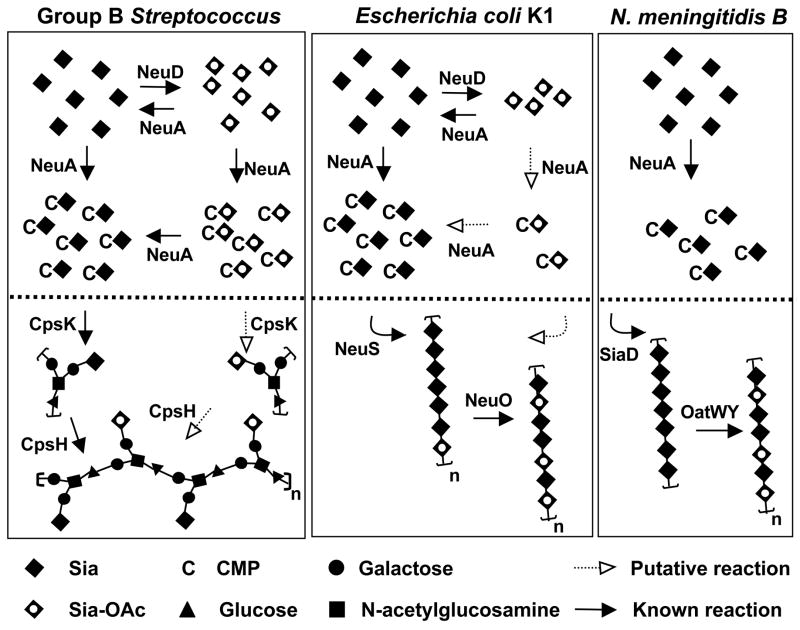
Figure 8. NeuA esterase activity in the context of capsule biosynthetic evolution in three prominent Sia-decorated pathogens GBS, E. coli K1, and serogroup B N. meningitidis.
Bacterial Sia biosynthesis can be divided into three main parts: 1) Sia biosynthesis and degradation, 2) intracellular Sia O-acetylation and de-O-acetylation, and 3) polysaccharide biosynthesis, polymerization, and O-acetylation. For simplicity, only 2) and 3) are shown (above and below the dotted line respectively). With respect to 1), these Sia-expressing pathogens use a homologous 3-enzyme pathway to generate N-acetylmannosamine (ManNAc) by epimerization of UDP-N-acetylglucosamine, condense phosphoenolpyruvate with ManNAc to generate N-acetylneuraminic acid (Neu5Ac), then activate Neu5Ac by transfer of the anomeric oxygen to the α-phosphate of CTP. Only E. coli possesses Sia degradation machinery. In GBS, cpsK and cpsH encode the sialyltransferase and repeating unit polymerase respectively. Intracellular Sia O-acetylation and de-O-acetylation is limited to GBS and E. coli, both of which possess the NeuD Sia O-acetyltransferase and NeuA Sia-O-acetyl esterase. The N. meningitidis pathway is more stream-lined in appearance due to the lack of an endogenous O-acetylation/de-O-acetylation cycle. Despite the pronounced difference between N. meningitidis (serogroup B) and E. coli K1 in intracellular O-acetylation cycling, these pathogens share an identical poly-Sia antigen made up of α2–8-linked Neu5Ac residues (polymerized by SiaD and NeuS respectively) that can be O-acetylated after capsule polymerization (by OatWY or NeuO respectively). See text for further discussion. Model is based on references (17, 18, 38, 51–61).