Abstract
Self-splicing of group I introns is accomplished by two sequential ester-transfer reactions mediated by sequential binding of two different guanosine ligands, but it is yet unclear how the binding is coordinated at a single G-binding site. Using a three-piece trans-splicing system derived from the Candida intron, we studied the effect of the prior GTP binding on the later ωG binding by assaying the ribozyme activity in the second reaction. We showed that adding GTP simultaneously with and prior to the esterified ωG in a substrate strongly accelerated the second reaction, suggesting that the early binding of GTP facilitates the subsequent binding of ωG. GTP-mediated facilitation requires C2 amino and C6 carbonyl groups on the Watson–Crick edge of the base but not the phosphate or sugar groups, suggesting that the base triple interactions between GTP and the binding site are important for the subsequent ωG binding. Strikingly, GTP binding loosens a few local structures of the ribozyme including that adjacent to the base triple, providing structural basis for a rapid exchange of ωG for bound GTP.
INTRODUCTION
Self-splicing of group I and group II introns and splicing of the eukaryotic pre-mRNA introns by spliceosome are all accomplished by two coupled ester-transfer reactions occurring at the 5′ and 3′ splice sites sequentially (1). However, it is not clear yet how the two ester-transfer reactions are coordinated. Extensive studies of the natural self-splicing and the engineered trans-acting group I introns have depicted many details of RNA structure and function (1–7), offering an appealing model system to address this question. The two ester-transfer reactions catalyzed by group I introns occur at a single guanosine-binding site (1,8). In the first reaction, an exogenous guanosine (exoG) binds to the active site and the 3′-hydroxyl group of this molecule attacks the 5′ splice site, leaving the 5′ exon with a free hydroxyl terminus. The exogenous guanosine then leaves the active site and the terminal guanosine residue (ωG) of the intron occupies it to initiate the second reaction. In this reaction, the free 3′-hydroxyl of the 5′ exon is aligned to attack the 3′ splice site, resulting in a released intron and ligated exons (1,9). The first reaction is commonly called cleavage, while the second is called exon ligation. One of the key issue in group I intron catalysis is how these two different guanosine factors are discriminated and how their bindings are coordinated by the intron during the first and second reactions.
It is apparent that conformational changes are required to switch the guanosine ligands between the first and second ester-transfer reactions, while the underlying mechanisms are not clear yet (10–12,10–12). Results from Tetrahymena ribozyme showed that the ribozyme structure is not pre-organized to readily bind the exoG ligand, instead, it is suggested that exoG binding requires a local but not a global structural rearrangement (13,14). After the completion of the cleavage reaction, binding affinity of exoG drastically deceases (15–17) and ωG readily exchanges for exoG to proceed to the exon ligation reaction. It has been shown that ωG effectively competes with exoG to occupy the active site, and therefore negatively regulates the ester-transfer reaction at the 5′ splice site (10,16,18). Interestingly, Zarrinkar and Sullenger have reported that exoG does not affect the exon ligation reaction, indicating that ωG binding is not competed by exoG (16). We noted that in their reaction system, ωG was physically attached to the ribozyme, therefore, the ribozyme folding and ωG binding events could not be dissected by the system.
To clear this hurdle, we developed a three-piece trans-splicing system in which the ribozyme folding, exoG binding and ωG binding can be distinguished. In this system, the Candida group I intron was truncated at the loop regions of P1 and P9.2; the 5′ and 3′ substrates that form P1 and P9.2-P9.0-ωG-P10, respectively, are supplied in trans. This design is very similar to what has been used to obtain the crystal structure of Azoarcus ribozyme (19). Some other groups have used three-piece systems of the Tetrahymnea ribozyme to investigate the kinetics of ωG binding. However, those systems usually contain a shorter ωG-containing 3′ substrate and none of them has ever been used to investigate how exoG modulates ωG binding (13,17,20,21). In contrast to the case of Tetrahymena ribozyme in which only 12% of the ribozyme folds to the catalytically active structure in the measurable time scale (22), the majority of the Candida ribozyme folds to the catalytically active structure through a relative fast-folding pathway (min–1) (23,24). Therefore, the Candida ribozyme gives us an opportunity to establish a functional link between the change of the catalytic activity and of the structure of the ribozyme upon GTP binding. This study demonstrated that exogenous GTP significantly facilitates the exon ligation reaction and that the facilitation is due to the prior binding of GTP to the ribozyme. Furthermore, we showed that the positive effect of GTP on ωG binding is accompanied by loosening the local ribozyme structure around the active site and at some peripheral regions. All these results point to the importance of the prior GTP binding in shaping a different ribozyme structure suitable for ωG binding.
MATERIALS AND METHODS
RNA preparation and nucleic acid labeling
The Ca.L-5 ribozyme (Figure 1A) was transcribed from the genomic DNA of Candida albicans and purified as described previously (23,25). RNA substrate of Ca/sub-6 (GCCUCU) was synthesized by Dharmacon (Lafayette, CO, USA). Ca/sub-7 (GCCUCUC) and ωG3′sub (GAUUUCUUGGAGCAAUGCUUAUG) were synthesized by Integrated DNA Technologies (Skokie, IL, USA). RNA substrates and the Ca.L-5 ribozyme for hydroxyl radical footprinting and native gel analysis were labeled at the 5′ end by the incorporation of [γ-32P] ATP (5000 Ci/mmol, Furui Biotech, Beijing) (23).
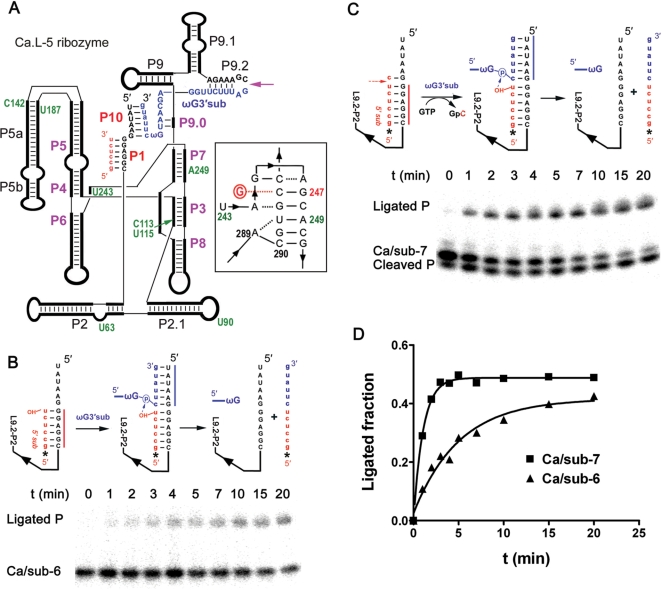
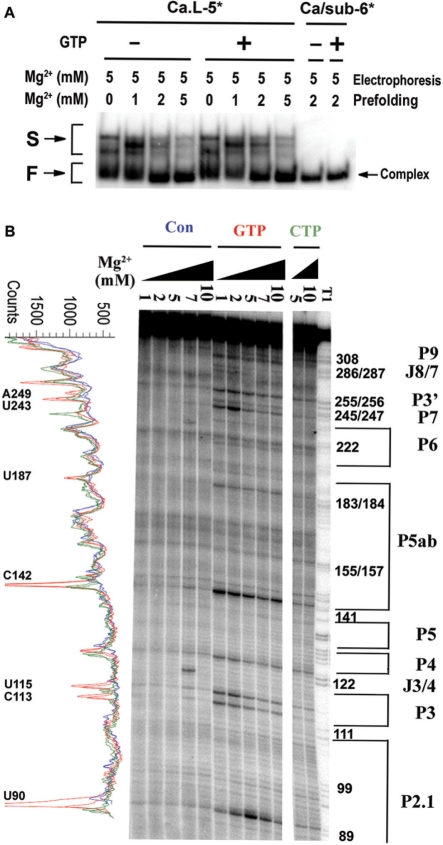
Figure 1.
A three-piece trans-splicing system derived from the Candida ribozyme. (A) Simplified secondary structure of the Ca.L-5 ribozyme. The thick black lines represent the ribozyme and the paired regions are labeled. The Ca.L-5 ribozyme is truncated at U6 in L1 and C362 in L9.2 (the purple arrow). Paired core structures of Ca.L-5 are labeled by purple bold letters while peripheral structures by black regular ones. Red lowercase letters refer to the 5′ substrate, which basepairs with the internal guide sequence of Ca.L-5 to form P1. The 3′ substrate (blue), named as ωG3′sub, forms P10–P9.0–P9.2 helixes; the lowercase letters refer to the 3′ exon. Positions indicated by green numbers refer to the sites that become more opened in the presence of GTP (Figure 5B). The proposed base triples encompassing the G-binding site are drawn according to the crystal structure of the Tetrahymena ribozyme (33) (insert on the right); circled red G refers to exoG. (B) Exon ligation reaction of Ca/sub-6. Ca.L-5 ribozyme (100 nM) was denatured and folded with [5′-32P] Ca/sub-6 (25 nM) at 37°C for 10 min. Then, the ligation reaction was initiated by adding ωG3′sub (250 nM). Upper panels illustrate the reaction process. The asterisk refers to the 5′ substrate radio-labeled at the 5′-end. Folding of Ca.L-5 ribozyme (black) with Ca/sub-6 (red) forms a ribozyme–substrate complex (left in the upper panel). ωG3′sub (blue) is added and binds to the complex to form an intermediate complex (middle in the upper panel). The intermediate complex undergoes the exon ligation reaction in which the 3′-hydroxyl group of Ca/sub-6 attacks the 3′ splice site in ωG3′sub, resulting in a free ribozyme and a ligated product (right in the upper panel). Lower panel is a representative gel of the exon ligation reaction of Ca/sub-6 at 3 mM MgCl2. Ligated P (12 nt) refers to the product of the ligation reaction. (C) Trans-splicing reaction of Ca/sub-7. The reaction was similarly performed as that of Ca/sub-6, except that Ca/sub-7 was used and 0.1 mM GTP was included. Upper panel illustrates the reaction process. Ca.L-5 (black) folds with Ca/sub-7 (red) to form a ribozyme–substrate complex (left in the upper panel). The complex undergoes the cleavage reaction when GTP is added and attacks the 5′ splice site in Ca/sub-7, resulting in a product of GpC. ωG3′sub (blue) binds to the complex and forms the same intermediate complex and final products (middle and right in the upper panel) as in (B). Lower panel is a representative gel of the trans-splicing reaction of Ca/sub-7 at 3 mM MgCl2. Cleaved P (6 nt) refers to the product from the cleavage at 5′ splice site, which is the same as Ca/sub-6; Ligated P (12 nt) refers to the product of the exon ligation reaction. (D) Quantification of the exon ligation reaction shown in (B) and (C). The ligation reactions were analyzed by calculating the fraction of Ligated P among the total substrate for Ca/sub-6 (black triangle) and the total cleaved product for Ca/sub-7 (black square), and then plotted against reaction time. The kobs for the exon ligation reaction of Ca/sub-7 and Ca/sub-6 were 0.94 and 0.17 min–1, respectively.
Analysis of the ribozyme activity
For the direct exon ligation reaction, the Ca.L-5 ribozyme diluted in 80 mM Tris–HCl was denatured and folded with Ca/sub-6 at 37°C for 10 min in 5 μl reaction containing 100 nM Ca.L-5 ribozyme, 50 mM Tris–HCl, 25 nM [5′-32P] Ca/sub-6, 250 nM ωG3′sub and the indicated concentrations of MgCl2 varying from 0 to 10 mM. ωG3′sub was added to initiate the reaction. For the cleavage-coupled exon ligation reaction, [5′-32P] Ca/sub-7 was used and 0.1 mM GTP was included. We found that lowering and increasing the concentration of Ca.L-5 ribozyme in the ligation reactions did not alter the equilibrium of the ligation reaction (Supplementary Figure S1).
To assay the effect of GTP on the exon ligation reaction, the reactions were performed with GTP added before or after folding the ribozyme with Ca/sub-6. As for the exon ligation reaction of GTP analogs, the Ca.L-5 ribozyme was folded in the presence of Ca/sub-6 and GTP analogs, and then the ligation rates were determined. ddGTP was purchased from Roche (Switzerland); [α-S] GTP, ITP, and 6′-thio GTP, were synthesized by Jena Bioscience GmbH (German).
To determine (kcat/Km)GTP, the Ca.L-5 ribozyme (200 nM) was denatured and folded with 32P-labeled substrates (12 nM) in the presence of 5 mM Mg2+ at 37°C for 10 min. Varying concentrations of GTP were added to initiate the cleavage reaction. kobs of each time-dependent reaction versus [GTP] was fitted to the Michaelis–Menten equation kobs = kcat × [GTP]/(Km + [GTP]). kcat and Km for ωG3′sub was similarly determined.
Footprinting analysis of the ribozyme structure
The Ca.L-5 ribozyme labeled at the 5′ end with [γ-32P] ATP was used for hydroxyl footprinting analysis as previously described (25). The 5′-end labeled RNA (∼20 ng) in 80 mM sodium cacodylate (pH 7) and 0.1 mM EDTA was denatured at 95°C for 1 min, annealed at 37°C for 3 min and then chilled on ice. Each sample was then folded at 37°C for 10 min after the addition of folding buffer containing 25 nM Ca/sub-6 and MgCl2 with desired concentrations. The hydroxyl radical footprinting was performed as previously described (25).
Native gel analysis of the ribozyme structure
The 5′-end labeled Ca.L-5 ribozyme was denatured at 95°C for 1 min, folded at 37°C for 10 min after the addition of folding buffer containing 25 nM Ca/sub-6 and MgCl2 with desired concentrations. All samples were chilled on ice and an equal volume of the loading dye containing 50% glycerol was added, followed by electrophoresis on a 5% native polyacrylamide gel (5 mM MgCl2) in 1×TB buffer (0.09 M Trisborate at pH 7.5 and 5 mM MgCl2).
RESULTS
A three-piece trans-splicing system of the Candida ribozyme
Ca.L-5 ribozyme was constructed to dissect the ribozyme folding, exoG binding and ωG-binding events. Ca.L-5 ribozyme is a truncated form of the Candida intron (23) that lacks the first five nucleotides in L1 and the last 17 nucleotides including ωG (Figure 1A). The 5′ and 3′ substrates that form P1 and P9.2-P9.0-ωG-P10, respectively, are provided in trans. Ca.L-5 catalyzes either a direct exon ligation reaction or a cleavage-coupled exon ligation reaction, depending on what kind of 5′ substrates are provided (Figure 1B and C). Footprinting analysis using Fe(II)-EDTA showed that the structure of Ca.L-5 ribozyme is similar to that of the ribozyme with an intact 3′-end (23, below), consistent with the finding that deletion of P9.2 and P9.0 has a marginal effect on the Mg2+-dependent folding of the Candida ribozyme (Unpublished data, P. Bao and Y. Zhang).
The 5′ substrate (Ca/sub-6) consists of six 5′-exonic nucleotides (GCCUCU) complementary to the internal guide sequence of Ca.L-5. This substrate forms a perfect P1 stem and its free 3′-OH group is ready for the in-line nucleophilic attack of the phosphor atom at the 3′ splice site, leading to the direct exon ligation reaction (Figure 1B, upper panel). Our previous study has revealed that the binding of the 5′ substrate is a rate-limiting step (26). To eliminate the binding complication, the Ca.L-5 ribozyme was folded with Ca/sub-6 at 37°C for 10 min prior to the exon ligation reaction. The 3′ substrate of ωG3′sub was then added to initiate the ligation reaction, and the predicted product of ligation was evident (Figure 1B, lower panel).
However, as shown in Figure 1C, when the Ca.L-5 ribozyme was pre-folded with a different 5′ substrate, Ca/sub-7 containing an additional nucleotide from the first position of the intron, both cleaved and ligated products were generated in the presence of GTP. The cleaved product was the same in size as Ca/sub-6, which was expected from the GTP-mediated precise cleavage at the 5′ splice site. The ligated product was exactly the same as that in the direct exon ligation reaction, indicating that only the Ca/sub-6 product resulting from the cleavage of Ca/sub-7, but not Ca/sub-7 itself, could initiate the exon ligation at the 3′ splice site, very possibly because the free 3′-OH group of Ca/sub-7 is not in-line for the nucleophilic attack.
Clearly, folding of the core and most peripheral interactions of the ribozyme, as well as formation of P1, could be separated from exoG and ωG binding in this three-piece system. By appropriately adjusting the order of addition of GTP and ωG, the effect of GTP in the exon ligation reaction as well as in coordinating splicing reactions could be determined.
The prior cleavage reaction at the 5′ splice site accelerates the following exon ligation at the 3′ splice site
We noted that both the cleavage and ligation reactions proceeded very fast when they were coupled. The significant accumulation of products from both reactions was evident in 1 min, the earliest time point of the experiments (Figure 1C). Surprisingly, the ligated product accumulated more slowly in the direct exon ligation reaction than in the cleavage-coupled one (Figure 1D), suggesting that the prior cleavage reaction at the 5′ splice site facilitates the following exon ligation at the 3′ splice site.
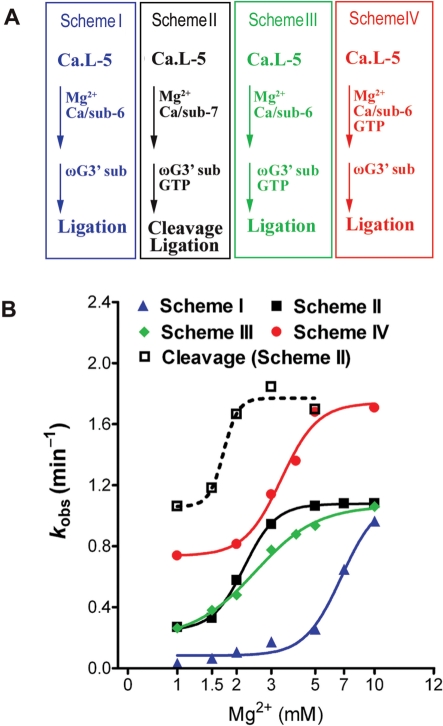
To detail this interesting observation, we obtained the observed first-order rates of the direct and cleavage-coupled exon ligation reactions under varying concentrations of magnesium. In the direct ligation reaction (Scheme I in Figure 2A), the ligation activity of Ca.L-5 increased slowly with low concentrations of magnesium, from 0.03 min−1 at 1 mM Mg2+ to 0.17 min−1 at 3 mM Mg2+ (Figure 2B). The ligation reaction was greatly facilitated by high concentrations of magnesium as demonstrated by the sharp increased kobs at Mg2+ > 5 mM, and the increase reached 0.96 min−1 at 10 mM Mg2+ (Figure 2B). The Mg2+ required for half-maximal catalytic activity (Mg1/2) was 6.77 mM, with a Hill coefficient of 4.7 (Table 1).
Figure 2.
Facilitation of the exon ligation at 3′ splice site by GTP. (A) Experiment procedures of the exon ligation reaction with or without GTP. Scheme I (blue) represents the direct exon ligation reaction where the ribozyme initially folded with Ca/sub-6. Scheme II (black) represents the cleavage-coupled exon ligation reaction where GTP was present and the ribozyme initially folded with Ca/sub-7, and the trans-splicing reaction occurred in this scheme. Scheme III (green) and IV (red) represent the GTP-facilitated ligation reactions. Scheme II is the same to Scheme I except for the simultaneous addition of 0.1 mM GTP and ωG3′sub in the reaction. Scheme IV differs from Scheme III in that GTP was added during the ribozyme folding. (B) The cleavage and exon ligation activities of Ca.L-5 ribozyme from four Schemes were plotted as described in Figure 1D. Same color was used to represent Schemes in (A). Dashed line refers to the cleavage of Scheme II. The fraction of Ca/sub-7 being cleaved by the ribozyme among the total substrate was calculated and plotted similarly as of the ligation product. kobs of each time-dependent reaction versus Mg2+ concentration was fitted to the Hill equation kobs = kmax × [Mg2+]n/(Mg1/2n + [Mg2+]n).
Table 1.
Magnesium-dependent catalysis by Ca.L-5 ribozyme
| Schemesa | Hillslope | Mg1/2 (mM) | |
|---|---|---|---|
| Cleavage | II | 11.7 | 1.72 |
| Exon ligation | I | 4.7 | 6.77 |
| II | 5.4 | 2.18 | |
| III | 2.8 | 2.48 | |
| IV | 4.7 | 3.36 |
aThe data from Figure 2 were plotted and summarized here.
In the cleavage-coupled exon ligation reaction (Scheme II in Figure 2A), both the cleavage and ligation activities of the Ca.L-5 ribozyme increased quickly at low concentrations of magnesium, and the cleavage reaction was catalyzed much faster than the ligation reaction (Figure 2B). The cleavage reaction reached the maximal activity (1.8 min−1) at 2–3 mM Mg2+, while the ligation reaction reached the maximal activity (1.1 min−1) at 3–5 mM Mg2+ (Figure 2B). These results suggested that the exon ligation reaction is rate-limiting in the coupled splicing reactions. Similar results were obtained when the natural Candida ribozyme was self-spliced (Supplementary Figure S2).
Comparison of the cleavage-coupled and direct exon ligation reactions under physiological magnesium concentrations (1–5 mM) revealed that exon ligation catalyzed by Ca.L-5 was much more efficient when exon ligation was coupled with cleavage (Figure 2B). For example, the catalytic rate was 0.27 min−1 at 1 mM Mg2+ in the coupled ligation reaction, which was 9-fold faster than that in the uncoupled reaction (0.03 min−1 at 1 mM Mg2+). When the natural Candida ribozyme underwent the self-splicing reaction, in which reactions at the 5′ and 3′ splice sites are coupled, the ligation activity was much higher than that of the two-piece ligation reaction (Supplementary Figures S2 and S3). Therefore, the finding that the cleavage reaction effectively accelerates the following exon ligation reaction can be biologically relevant.
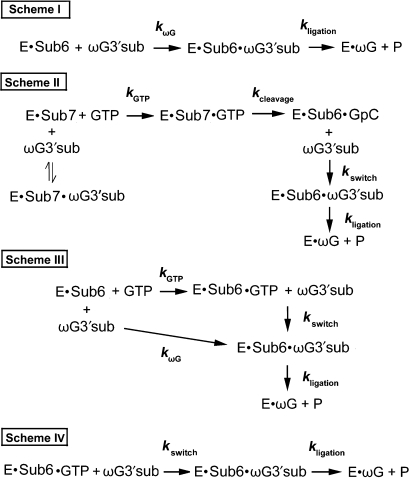
In order to distinguish the possible contributions of different kinetics to the observed kobs, we delineated the cleavage-coupled and direct exon ligation reactions in Figure 3. As shown in Table 2 and Supplementary Figure 4, the chemical steps of the cleavage reaction and the exon ligation reaction were faster than the corresponding kobs. Therefore, we reasoned that kobs reflects the binding rate of GTP or ωG. The cleavage activity of Ca.L-5 was higher than the ligation activity in the coupled reaction, suggesting that kGTP > kswitch. kswitch refers to the exchange between GTP and ωG at the G-binding site. Compared to the cleavage-coupled ligation reaction, the direct exon ligation reaction occurred at a much lower rate constant. This result pointed out that switching of GTP to ωG at the G-binding site is faster than the direct binding of ωG, i.e. kswitch > kωG.
Figure 3.
Illustration of kinetic steps of exon ligation in all the reactions listed in Figure 2. E refers to Ca.L-5 ribozyme. Sub6 and Sub7 refer to Ca/sub-6 and Ca/sub-7, respectively. In Scheme I, E·Sub6 represents the complex initially formed during ribozyme folding. ωG3′sub binding to this complex forms E·Sub6·ωG3′sub complex at a rate constant of kωG. The three-piece complex performs the exon ligation reaction at a rate constant of kligation. In Scheme II, E·Sub7 complex was formed during ribozyme folding. And the subsequent GTP binding to the complex forms E·Sub7·GTP (kGTP), which is followed by the cleavage reaction at the 5′ splice site (kcleavage). The resulted E·Sub6·GpC complex is bound by ωG3′sub, and then ωG switches GpC from the G-binding site (kswitch), resulting in the E·Sub6·ωG3′sub complex that undergoes the ligation reaction (kligation). ωG3′sub can also binds to E·Sub7 to form E·Sub7·ωG3′sub complex, but this complex is inactive and unable to undergo the splicing events. In Scheme III, two pathways of exon ligation are shown. E·Sub6 complex can bind to ωG3′sub and initiate the ligation pathway the same as that in Scheme I. Alternatively, GTP binds E·Sub6 and forms E·Sub6·GTP. Then, ωG3′sub binds to E·Sub6·GTP, and ωG switches GTP from the G-binding site (kswitch), resulting in the E·Sub6·ωG3′sub undergoing the ligation reaction (kligation). In Scheme IV, E, GTP and Sub6 initially form E·Sub6·GTP complex, the binding of ωG3′sub allows ωG to switch GTP from the G-binding site (kswitch), resulting in the E·Sub6·ωG3′sub complex to undergo exon ligation (kligation).
Table 2.
The kinetic parameters of Ca.L-5 ribozyme
| GTP | ωG3′sub | |
|---|---|---|
| kcat/Km (M−1 min−1) | 2.0 × 106 | 6.0 × 104 |
| Km (μM) | 3.2 | 81.2 |
| kcat (min−1) | 6.4 | 4.9 |
Note: The kcat, Km and kcat/Km were determined as described in ‘Materials and Methods’ section at 37°C, 5 mM MgCl2.
GTP binding facilitates ωG binding
We then wanted to address why kswitch > kωG. The cleavage-coupled ligation reaction (Scheme II) differs from the direct ligation reaction (Scheme I) in three aspects including the presence of GTP, an extra nucleotide and the cleavage reaction at the 5′ splice site. Ca/sub-6 bound to Ca.L-5 ribozyme with an affinity much higher than Ca/sub-7, excluding the possibility that a lower binding affinity of Ca/sub-6 accounts for the small kωG (Supplementary Figure 5). Using the three-piece system, the effect of exoG on exon ligation at the 3′ splice site could be studied by adding GTP to the direct exon ligation reaction (Scheme III in Figure 2A), which bypasses the complication of the other two factors. As shown in Figure 2B, the presence of GTP in the direct ligation reaction greatly promoted the ligation activity of Ca.L-5, and the extent of promotion was almost identical to that coupled with cleavage. These results suggested that it is the GTP binding in the cleavage-coupled reaction that promotes the subsequent ωG binding for exon ligation. It is noticeable that when GTP was added in the direct exon ligation reaction, ωG could reach the G-binding site through two independent pathways, i.e. switching of GTP or direct binding (Figure 3, Scheme III). This explained why the ligation activity in the GTP-facilitated ligation reaction (Scheme III) was less cooperative with respect to magnesium than that in the cleavage-coupled reaction (Scheme II) where only the coupled ωG binding was effective (Table 1).
We then added GTP during the ribozyme folding prior to the addition of ωG3′sub, so that GTP may maximally occupy the G-binding site and highly synchronize the formation of ribozyme–substrate–GTP complex without the competition by ωG (Scheme IV in Figures 2 and 3). Strikingly, the ligation activity of this reaction was significantly higher than those of the cleavage-coupled and GTP-facilitated reactions (Figure 2B). For example, the catalytic rate was 0.74 min−1 at 1 mM Mg2+ and reached 1.71 min−1 at 10 mM Mg2+. This finding indicated that the pre-folded ribozyme–substrate–GTP complex (Scheme IV) may differ from the pre-folded ribozyme–substrate complex (Scheme III) in structure, with the former one binding ωG more readily than the latter.
All these results are consistent with a conclusion that the early binding of GTP to the ribozyme shapes a more favorable local environment at the active site to accommodate ωG.
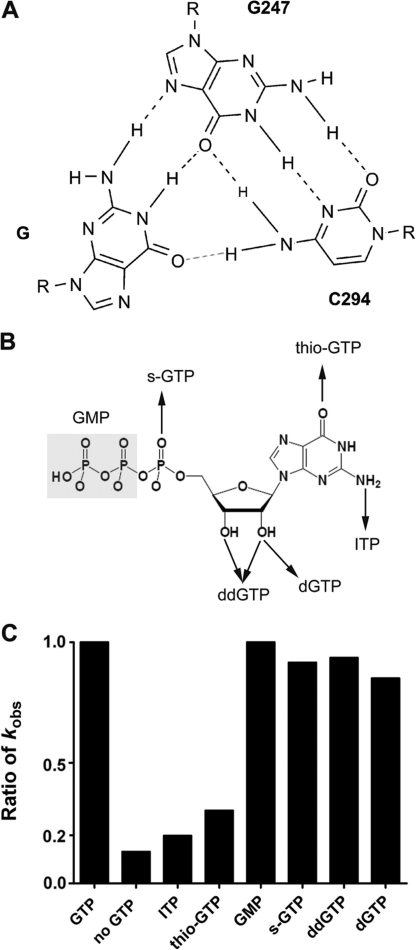
GTP facilitates ωG binding through forming a base triple at the G-binding site
Then, we wanted to explore how GTP facilitates ωG binding by addressing which chemical group of GTP is important in the facilitation of ωG binding. It has been revealed that a guanosine ligand binds to the G-binding site of group I introns via a base triple (Figure 4A) (8,27). GTP analogs with different substitutions of chemical groups were used in this study, with two of them containing chemical modifications at the 2-amino group (ITP) and 6-carbonyl group (thio-GTP) that participate in the formation of the G·C-G base triple (Figure 4B). Exon ligation reactions with GTP analogs were performed as in Scheme IV of Figure 2A. Ratios of the ligation kobs with GTP analogs to that with GTP were obtained and defined as the relative activity (Figure 4C).
Figure 4.
GTP binding facilitates ωG binding through forming a base triple at the G-binding site. (A) Base-triple interactions between guanosine and the active site of the Candida ribozyme. G247 and C294 locate at P7. R refers to ribose of the nucleotide. Hydrogen bonds between N2 of GTP and N7 of G247, N1 of GTP and O6 of G247 have been proven by Michel et al. (27). The hydrogen bond between O6 of GTP and N4 of C294 (red dashed line) is proposed based on its importance in promoting ωG binding (this study) and in supporting the self-splicing reaction (29). All three hydrogen bonds are formed between ωG and the binding site in the crystal structure of Tetrahymena ribozyme (33). (B) Simplified diagrams of guanosine analogs. (C) Exon ligation activities in the presence of GTP analogs. The reaction was performed as in Scheme IV in Figure 2 in the presence of 2 mM Mg2+. Ratio of kobs was obtained by dividing the ligation activity of GTP analog by that of GTP. No GTP refers to the exon ligation reaction in the absence of GTP.
Strikingly, the chemical modifications at both C6 and C2 positions of GTP dramatically reduced the effect of GTP in stimulating the ligation rate of Ca.L-5, and the reduction was about 3-fold and 5-fold lower for thio-GTP and C2 amine (inosine), respectively. The accumulated reduction at these two positions was about the same as the GTP-lacking sample, suggesting the interactions between these two groups and the G-binding site are the major requirement to stimulate the following ωG binding.
Consistently, stimulation of the ligation activity by GTP was retained for all the other analogs, including those with the oxygen atom of the α-phosphate being replaced by a thio (s-GTP), or those lacking any one of the following: β- and γ-phosphates (GMP), 2′-OH and/or 3′-OH groups (ddGTP, dGTP). Consistently, the unimportance of phosphates in GTP binding to group I ribozyme has been reported previously (1,28,29). However, the 2′- and 3′-OH groups, which are indispensable for the cleavage reaction as well as the self-splicing reaction (17,29–31), are not required for promoting ωG binding, supporting the conclusion that the ester-transfer reaction at the 5′ splice site is not required for stimulating ωG binding.
These results strongly suggested that the facilitation of ωG binding by GTP binding is due to their same base-triple interactions with the G-binding site, leading to an elegant mechanism for GTP to shape a more favorable binding site for ωG.
GTP binding loosens a few local structures
Native PAGE gel electrophoresis was used to detect whether the global fold of Ca.L-5 ribozyme was changed by GTP binding. As shown in Figure 5A, the Ca.L-5 ribozyme was pre-folded in the presence of the substrate of Ca/sub-6 and varying concentrations of Mg2+, and then subjected to electrophoresis on native PAGE gels containing 5 mM Mg2+. Both fast migrated and slowly migrated ribozyme species were evident when Ca.L-5 was radio-labeled. When the substrate was radio-labeled, only the fast-migrated band appeared, suggesting that this band contains the substrate-bound ribozyme population. Interestingly, addition of GTP did not alter the distribution of different ribozyme population nor introduce any new conformation, consistent with a model that the ribozyme may adopt a similar fold when GTP occupies the binding site.
Figure 5.
GTP binding to the ribozyme induces a looser core structure. (A) Native gel electrophoresis of the folded Ca.L-5 ribozyme with (+) or without GTP (–). Both the native polyacrylamide gel and the TB buffer contained 5 mM MgCl2. The asterisk referred the radio-labeled RNA species. F and S refer to fast and slow-migrated ribozyme species, and complex represents the ribozyme bound by Ca/sub-6. (B) Hydroxyl radical footprinting analysis of the tertiary structures of Ca.L-5 in the presence and absence of 0.1 mM GTP and varying concentrations of magnesium. Hydroxyl radical footprinting was performed as described in ‘Materials and Methods’ section. The band intensity profiles of lanes with 5 mM and 10 mM Mg2+ on the footprinting gels were obtained as described previously (23) and shown in the left. Opened regions of the Ca.L-5 ribozyme in the presence of GTP are labeled. Red and brown refer to the ribozyme folding in the presence of GTP and CTP, respectively, while blue refers to that in the absence of both GTP and CTP (Con).
In order to detect the ribozyme structure upon GTP binding, Fe(II)-EDTA footprinting experiments were performed. Please be noted that substrate was also included in these experiments as in all other experiments. CTP was used as a control to eliminate the non-specific action of nucleotides on the ribozyme structure (Figures 1A and 5B). In the presence and absence of GTP, magnesium-dependent protection of the ribozyme back bone was observed at a number of regions, consistent with the magnesium-dependent folding of the tertiary structure of the ribozyme. Interestingly, GTP did not change the hydroxyl radical protection pattern of the ribozyme, but rather makes certain nucleotides much more prone to cleavage. This is consistent with the conclusion that GTP binding does not change the global fold of the ribozyme, but rather changes a few local structures.
We have noticed that CTP slightly increases the accessibility of a few positions of the ribozyme to the hydroxyl radical. Therefore, only positions significantly more accessible in the presence of GTP than those of CTP have been considered as the GTP-opened sites. These sites included U243 and A249 that are close to the base-triple interaction formed between GTP and the binding site, suggesting that GTP binding significantly loosens the local structure around its binding site. Moreover, C113 and U115 in P3, U90 in P2.1, C142 in P5a and U63 in P2 (Figure 5B and data not shown) all became more accessible to the free radical, showing that in addition to the G-binding site region, some other regions of the core structure (P3) and peripheral structures (P2, P2.1 and P5a) are somewhat relaxed by GTP binding as well. These results suggest that GTP binding results in a significant change of a few local structures of the ribozyme including the G-binding site. This change provides structural basis for a more suitable and easily accessible pocket for ωG binding.
DISCUSSION
Facilitation of ωG binding by exoG: ωG binds slow or fast?
It is well known that only one guanosine-binding site is present in the group I intron ribozyme, while two guanosine ligands sequentially occupy the site during the intron splicing reaction. It has been found that ωG, the latter bound ligand, can inhibit the binding of the early ligand exoG to the active site, while the exoG does not compete with the tightly bound ωG (16,18). In this study, using the three-piece trans-splicing model derived from the Candida ribozyme, we showed that exoG significantly accelerates ωG binding by forming a base-triple interaction with the G-binding site. This finding reveals that the sequential binding of these two guanosine ligands to the single active site confers functional importance in group I intron splicing.
However, facilitation of ωG binding by exoG has not been uncovered by the study of the Tetrahymena ribozyme (16), which could be due to the use of two-piece trans-splicing model. In that system, ωG is physically attached to the ribozyme, and the ribozyme folding and ωG binding events cannot be dissected. If ωG accesses the G-binding site during the ribozyme folding, the latter added exoG cannot compete with the pre-bound ωG.
We have demonstrated that binding of the trans-supplied ωG substrate is much slower than binding of the free GTP. This slow binding could be attributed to the low affinity of the ωG substrate to the ribozyme, which is about 20-fold lower than that of GTP (Table 2). The ωG substrate can form P9.0 and P10 with the Ca.L-5 ribozyme, and formation of P9.0 and P10 drastically enhances the affinity of binding guanosine substrates to the Tetrahymena ribozyme (13). Strikingly, binding affinity of a similar ωG substrate to the Tetrahymena ribozyme is much higher than that of the Candida ribozyme, suggesting that the detailed mechanism of ωG binding may differ between these two ribozymes which belong to different subgroups.
On the other hand, previous study of the Tetrahymena ribozyme showed that binding affinity of the esterified ωG substrate is drastically lower than that of the substrate having a 3′-OH end (17). Consistently, we found that binding of the ωG present at the 3′ terminal end of the Candida ribozyme is at the similar rate with that of GTP, in the aspect of catalyzing the first step reaction (Supplementary Figure S6). In our three-piece trans-splicing system, ωG is esterified and no 3′ hydroxyl group is present. ωG is also structurally constrained because of the presence of P9.0–P9.2 and P10 at the upstream and downstream, respectively. This structurally constrained and esterified ωG resembles the natural context, and is analogous to the one used to obtain the high-resolution structure of the Azoarcus ribozyme (14,19). Crystal structures of several group I introns showed that the ωG-C:G base triple is sandwiched by three other base triples in the catalytic core (14,19,32,33), suggesting that forming the correct interactions between the esterified ωG and the G-binding site is not structurally favorable. The slow ωG binding is readily explained as that the initially folded ribozyme structure does not favor the binding of the constrained ωG to the active site.
Binding of exoG induces openness at a few local structures
Study of the Tetrahymena ribozyme suggested that G-binding site is not pre-organized, but is induced upon GTP binding (13,34). Structural analysis of the Azoarcus ribozyme predicted the disruption of local ribozyme structures for the exchange of ωG and exoG during the two-step ester-transfer reactions (11,14). Here, we provided direct evidence that binding of exoG to the folded ribozyme remarkably alters the ribozyme structure by loosening a number of structurally important positions, suggesting the disruption of some tertiary interactions. Among the opened positions, two are located at two sides of the three juxtaposed base triples including the GTP base triple (Figure 1A). Openness at position U243 is very interesting because U243 forms base triple with P4, which is critical for closing the P4–P6 and P3–P7 core domains. The opened A249 is also striking because the base pair between A249 and U292 is a predicted donor of A289 in forming a base triple. Clearly, the increased openness at these two positions indicated the reduced tertiary interactions, which likely shapes local environments more favorable for ωG binding. In addition, the altered structure is less compacted at the core region P3 and several peripheral regions including P5a, P2 and P2.1, indicating that the structure changes are more spread than expected.
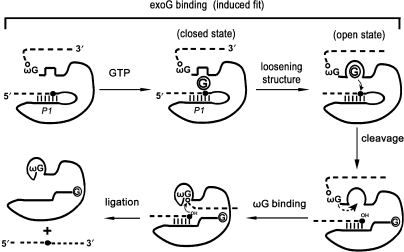
In summary, the increased structural flexibility around the G-binding site upon GTP binding provides structural basis for the easier access of the structurally constraint ωG. Breaking interactions at other regions may also contribute to the easier docking of ωG. In this way, the prior GTP binding prepares the ribozyme structure for the subsequent ωG binding and thus promotes the exon ligation at the 3′ splice site (Figure 6).
Figure 6.
GTP binding converts the ribozyme to an open state competent for ωG binding. Dashed lines and solid lines represent exons and introns, respectively. The base-paired P1 helix is shown. Solid black dot indicates the 5′ splice site while open dot indicates the 3′ splice site. Circled G refers to exogenous GTP. The induced-fit binding of guanosine to the Tetrahymena ribozyme has been previously reported (13).
The importance of forming base-triple interactions between exoG and the G-binding site: how far away from ωG binding
Interestingly, C2 amino group and C6 carbonyl group at exoG are essential for the acceleration of ωG binding. The corresponding positions of C2 and C6 on ωG participate in the base-triple interactions between ωG and G-binding site, as revealed by crystal structures of the Twort and Tetrahymena ribozymes (32,33). Therefore, we propose that formation of the base triple with G-binding site is essential for inducing and perhaps stabilizing the opened local structure of the group I ribozyme. Once the G-binding site is correctly folded, exoG and ωG can exchange much more rapidly. In addition, we found that the second step of splicing is still less efficient than the first one in the presence of GTP (Figures 1C and 2 Scheme II), suggesting additional structural transition after the first reaction (10,11).
Our results and previous studies support a model illustrating the mechanism underlining the facilitation of ωG binding by exoG (Figure 6) (13,15–17,34). We propose that when it is present, the free exoG first encounters the closed active site, and then through an induced-fit conformational change process, the correct base-triple interactions forms between exoG and the binding site. On the other hand, the closed active site disfavors binding of the structurally constrained and esterified ωG in the precursor RNA. The preferred binding of GTP initiates the first step reaction and prevents the non-productive reaction mediated by ωG. After the first step reaction at the 5′ splice site, exoG is esterified to the 5′-end of the intron and becomes structurally constrained. Meanwhile, exoG binding converts the ribozyme from the closed state into the open one, which allows the structurally constrained ωG to exchange for the reacted exoG rapidly. The accomplishment of the first reaction also provides a correctly positioned P1 with a free 3′-OH that coordinates ωG binding (19). At last, successful binding of ωG initiates the second step reaction at the 3′ splice site and the splicing is completed. This model may provide some insights into the mechanisms of coordinating two sequential reactions at the 5′ and 3′ splice sites by group II introns and spliceosomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (grant 30330170 to Y.Z.); National Basic Research Program (973) of China (grant 2005CB724604 to Y.Z.). Funding for open access charge: National Basic Research Program (973) of China.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. David M.J. Lilley (University of Dundee) for his helpful suggestion.
REFERENCES
- 1.Cech TR. Self-splicing of group I introns. Annu. Rev. Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 3.Narlikar GJ, Herschlag D. Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes. Annu. Rev. Biochem. 1997;66:19–59. doi: 10.1146/annurev.biochem.66.1.19. [DOI] [PubMed] [Google Scholar]
- 4.Thirumalai D, Lee N, Woodson SA, Klimov D. Early events in RNA folding. Annu. Rev. Phys. Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 5.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 6.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Hougland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D. In: The RNA World. 3rd. Gesteland RF, Cech TR, Atkins JF, editors. New York: Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 8.Been MD, Perrotta AT. Group I intron self-splicing with adenosine: evidence for a single nucleoside-binding site. Science. 1991;252:434–437. doi: 10.1126/science.2017681. [DOI] [PubMed] [Google Scholar]
- 9.Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem. Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Golden BL, Cech TR. Conformational switches involved in orchestrating the successive steps of group I RNA splicing. Biochemistry. 1996;35:3754–3763. doi: 10.1021/bi952599z. [DOI] [PubMed] [Google Scholar]
- 11.Rangan P, Masquida B, Westhof E, Woodson SA. Architecture and folding mechanism of the Azoarcus Group I Pre-tRNA. J. Mol. Biol. 2004;339:41–51. doi: 10.1016/j.jmb.2004.03.059. [DOI] [PubMed] [Google Scholar]
- 12.Emerick VL, Pan J, Woodson SA. Analysis of rate-determining conformational changes during self-splicing of the Tetrahymena intron. Biochemistry. 1996;35:13469–13477. doi: 10.1021/bi960865i. [DOI] [PubMed] [Google Scholar]
- 13.Karbstein K, Herschlag D. Extraordinarily slow binding of guanosine to the Tetrahymena group I ribozyme: implications for RNA preorganization and function. Proc. Natl Acad. Sci. USA. 2003;100:2300–2305. doi: 10.1073/pnas.252749799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McConnell TS, Cech TR, Herschlag D. Guanosine binding to the Tetrahymena ribozyme: thermodynamic coupling with oligonucleotide binding. Proc. Natl Acad. Sci. USA. 1993;90:8362–8366. doi: 10.1073/pnas.90.18.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zarrinkar PP, Sullenger BA. Probing the interplay between the two steps of group I intron splicing: competition of exogenous guanosine with omega G. Biochemistry. 1998;37:18056–18063. doi: 10.1021/bi982193x. [DOI] [PubMed] [Google Scholar]
- 17.Karbstein K, Carroll KS, Herschlag D. Probing the Tetrahymena group I ribozyme reaction in both directions. Biochemistry. 2002;41:11171–11183. doi: 10.1021/bi0202631. [DOI] [PubMed] [Google Scholar]
- 18.Mei R, Herschlag D. Mechanistic investigations of a ribozyme derived from the Tetrahymena group I intron: insights into catalysis and the second step of self-splicing. Biochemistry. 1996;35:5796–5809. doi: 10.1021/bi9527653. [DOI] [PubMed] [Google Scholar]
- 19.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 20.Bevilacqua PC, Sugimoto N, Turner DH. A mechanistic framework for the second step of splicing catalyzed by the Tetrahymena ribozyme. Biochemistry. 1996;35:648–658. doi: 10.1021/bi951962z. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Turner DH. Effects of Mg2+ and the 2′ OH of guanosine on steps required for substrate binding and reactivity with the Tetrahymena ribozyme reveal several local folding transitions. Biochemistry. 1997;36:11131–11139. doi: 10.1021/bi971034v. [DOI] [PubMed] [Google Scholar]
- 22.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 23.Xiao M, Leibowitz MJ, Zhang Y. Concerted folding of a Candida ribozyme into the catalytically active structure posterior to a rapid RNA compaction. Nucleic Acids Res. 2003;31:3901–3908. doi: 10.1093/nar/gkg455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Xiao M, Lu C, Zhang Y. Fast formation of the P3-P7 pseudoknot: a strategy for efficient folding of the catalytically active ribozyme. RNA. 2005;11:59–69. doi: 10.1261/rna.7145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao M, Li T, Yuan X, Shang Y, Wang F, Chen S, Zhang Y. A peripheral element assembles the compact core structure essential for group I intron self-splicing. Nucleic Acids Res. 2005;33:4602–4611. doi: 10.1093/nar/gki770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang YF, Xiao M, Yin P, Zhang Y. Monovalent cations use multiple mechanisms to resolve ribozyme misfolding. RNA. 2006;12:561–566. doi: 10.1261/rna.2188306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michel F, Hanna M, Green R, Bartel DP, Szostak JW. The guanosine binding site of the Tetrahymena ribozyme. Nature. 1989;342:391–395. doi: 10.1038/342391a0. [DOI] [PubMed] [Google Scholar]
- 28.Tanner NK, Cech TR. Guanosine binding required for cyclization of the self-splicing intervening sequence ribonucleic acid from Tetrahymena thermophila. Biochemistry. 1987;26:3330–3340. doi: 10.1021/bi00386a013. [DOI] [PubMed] [Google Scholar]
- 29.Bass BL, Cech TR. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: implications for biological catalysis by RNA. Nature. 1984;308:820–826. doi: 10.1038/308820a0. [DOI] [PubMed] [Google Scholar]
- 30.Sjogren AS, Pettersson E, Sjoberg BM, Stromberg R. Metal ion interaction with cosubstrate in self-splicing of group I introns. Nucleic Acids Res. 1997;25:648–653. doi: 10.1093/nar/25.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinstein LB, Jones BC, Cosstick R, Cech TR. A second catalytic metal ion in group I ribozyme. Nature. 1997;388:805–808. doi: 10.1038/42076. [DOI] [PubMed] [Google Scholar]
- 32.Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- 33.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol. Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Profenno LA, Kierzek R, Testa SM, Turner DH. Guanosine binds to the Tetrahymena ribozyme in more than one step, and its 2′-OH and the nonbridging pro-Sp phosphoryl oxygen at the cleavage site are required for productive docking. Biochemistry. 1997;36:12477–12485. doi: 10.1021/bi9708895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.