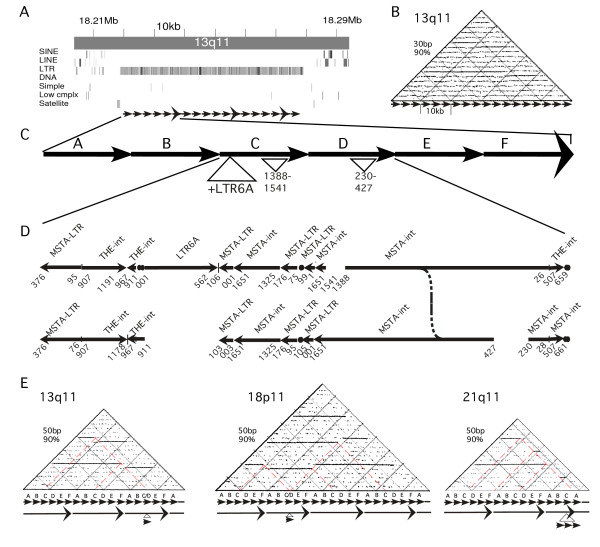
Figure 5.
Analysis of the repeat unit structure of the LTR arrays from chromosomes 13, 18 and 21. A) Genomic region from chromosome 13q11 containing the LTR array. The REPEAT MASKER Tracks from the UCSC genome browser are shown, which indicate the large 60 kb array of LTR transposons. Homologous monomeric repeat units are indicated by arrows. B) Self similarity dot plot of LTR array. 30 bp windows at 90% homology reveals the ~3.5 kb monomeric repeat units as horizontal lines. C) Schematic of a higher-order repeat unit (HOR) consisting of 6 ~3.5 kb monomeric repeat units. The insertion of an LTR6A into monomer C of each HOR is shown, as well as additional deletions of the MSTA-int repeats. D) Detail of the composition of monomers C and D indicating the MaLR LTR fragments that make up the repeat units, taken from the REPEAT MASKER output and numbered relative to the consensus for each element. The insertion of a full length LTR6A into monomer C can be seen. E) Self-similarity dot plots of LTR array from chromosome 13q11, 18p11 and 21q11 at 50 bp windows 90% homology. The HOR organization is revealed as bold solid horizontal lines, and are shown schematically by arrows below. Putative unequal crossing over events unique to each LTR array are revealed by the gaps and shift of these lines, and deleted monomeric repeat units indicated below.