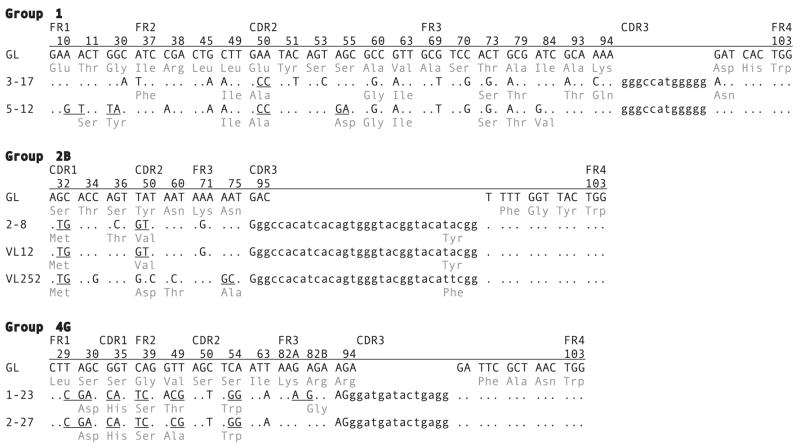
FIGURE 6.
Comparisons of intraclonally derived cDNA sequences. cDNA sequences were selected from shark-33 epigonal organ library after hybridization with probes to VH and CH. Group 1 cDNA sequences 3–17 and 5–12, which have the same CDR3, are compared with the reference G1 VH germline gene and JH flank. The codon, the codon positions, and the names of regions (FR1, CDR1, FR2, CDR2, FR3, CDR3, and FR4) are shown and compared with the substituted positions in the mutants. The portion of CDR3 consisting of N/P region and D segment sequences is indicated in lower case. Dots indicate identity with the germline, point mutations are shown, and tandem (contiguous) changes are indicated by underlining. For example, codons 10 and 11, GAA ACT, are changed to GAG TCT in cDNA 5–12. This change caused a replacement substitution in codon 11, which is shown in the three-letter amino acid code. In addition to CDR3, 3–17 shares eight point mutations and one tandem doublet with 5–12. Group 2B cDNA sequences 2–8, VL12, and VL252 share CDR3 and one doublet mutation. Whereas 2–8 differs from VL12 by one point mutation, both of them had diverged much earlier from VL252. Group 4G cDNA sequences 1–23 and 2–17 share CDR3, two point mutations and five tandem mutation sets.