FIGURE 1.
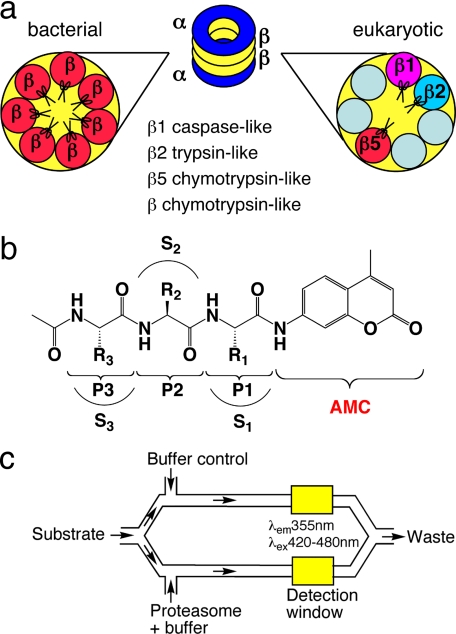
Schematic illustration of enzymes, substrates, and assay system. a, different composition of β subunits in eubacterial and eukaryotic proteasomes. The Mtb proteasome has seven identical β subunits, whereas eukaryotic proteasomes have seven different β subunits. Only three β subunits of eukaryotic proteasomes are active once their Thr1 active sites are exposed by autocatalytic removal of propeptide: β1, caspase-like; β2, trypsin-like; and β5, chymotrypsin-like. Eubacterial proteasomes are generally considered to be chymotrypsin-like when assayed with small peptide substrates. b, structure of the acetyl-P3-P2-P1-AMC substrate library. R1, R2, and R3 refers to the side chains of P1, P2, P3 amino acids, respectively; and S1, S2, and S3 refer to the binding pockets of the proteasome for P1, P2, and P3 amino acid side chains, respectively. c, schematic illustration of microfluidic TF460 assay system. Stream splitting and simultaneous fluorescence detection facilitated subtraction of the background fluorescence of each unhydrolyzed substrate; after substrate in buffer was pulled into the channel through vacuum, the stream was split 50:50 into parallel channels and mixed with enzyme and buffer, respectively. The final annotated data were obtained by subtracting background fluorescence of the unhydrolyzed substrate from that of the enzymatic reaction.