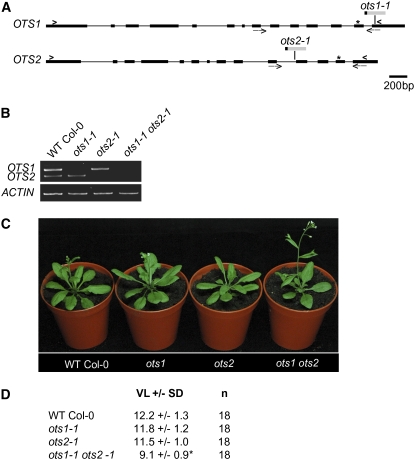
Figure 2.
OTS1 and OTS2 Act Redundantly to Affect Flowering Time.
(A) Schematic representation of the OTS1 and OTS2 genomic DNA regions. Exons are represented as black boxes connected by thin lines (introns), and arrowheads on each sequence represent the positions of the start and stop codons. Asterisks indicate the position of the Cys active site. T-DNA insertion sites are shown as gray boxes with the name of the corresponding allele. Black boxes at the extremities of the T-DNA show the location of the left border. Arrows below the genomic sequence represent the positions of the forward and reverse primers used for RT-PCR analysis.
(B) RT-PCR analysis of OTS1 and OTS2 transcript levels from total RNA derived from young seedlings of the indicated genotypes. OTS1 and OTS2 transcripts were amplified in parallel using the primers (arrows) indicated in (A) for 30 cycles of PCR, and ACTIN was used as a loading control. The experiment was repeated at least four times
(C) Phenotypes of representative 25-d-old ots1 (ots1-1), ots2 (ots2-1), and ots1 ots2 (ots1-1 ots2-1) plants grown in long-day conditions compared with wild-type Col-0.
(D) Quantification of flowering time as average number of vegetative leaves (VL) of the indicated genotypes with sd. The asterisk indicates a significant difference of flowering time from wild-type Col-0 (P < 0.01).