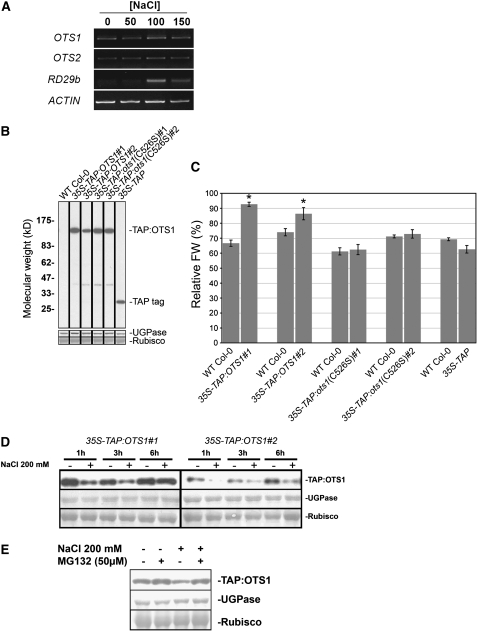
Figure 6.
OTS1 Protein Levels Are Downregulated by Salt.
(A) RT-PCR transcript analysis (25 cycles) of OTS1 and OTS2 levels from total cDNA derived from seedlings grown for 12 d on plates supplemented with the indicated salt concentrations. ACTIN served as a loading control, while RD29b, a gene known to be upregulated by salt, was used as a positive control. The RT-PCR analysis was repeated three times with similar results.
(B) Immunoblot with PAP (detects the protein A domain of the TAP tag) of 10 μg of total protein derived from independent transgenic lines ectopically expressing TAP:OTS1, TAP:ots1(C526S), and TAP tag alone and compared with wild-type Col-0. The bands corresponding to the TAP:OTS1 and TAP:ots1(C526S) fusion proteins or TAP tag are indicated at right. In the bottom panels, UGPase immunostaining and Ponceau staining of Rubisco large subunit were used as loading controls.
(C) Quantification of fresh weight (FW) on 100 mM NaCl–supplemented plates expressed as a percentage relative to the untreated condition in five independent transgenic lines, with each one being compared with the wild type. Error bars indicate se (n = 16 to 20). Asterisks indicate significant differences from the wild-type control (P < 0.01).
(D) Immunoblot with PAP of 10 μg of total proteins derived from two independent transgenic lines ectopically expressing TAP:OTS1. Seedlings were grown for 8 d in liquid 0.5× MS medium, after which they were treated with 200 mM NaCl (+) or distilled water (−) as a control and harvested at the indicated time points.
(E) Eight-day-old seedlings were incubated for 3 h with MG132 dissolved in DMSO or DMSO alone and subsequently treated with NaCl (200 mM) or distilled water as a control. Ten micrograms of total proteins was fractionated on an 8% SDS-PAGE gel, and TAP:OTS1 was detected with PAP. UGPase immunostaining and Ponceau staining of Rubisco large subunit were used as loading controls.