Figure 1. microRNA miR-132 and miR-219-1 are differentially regulated by CREB and CLOCK.
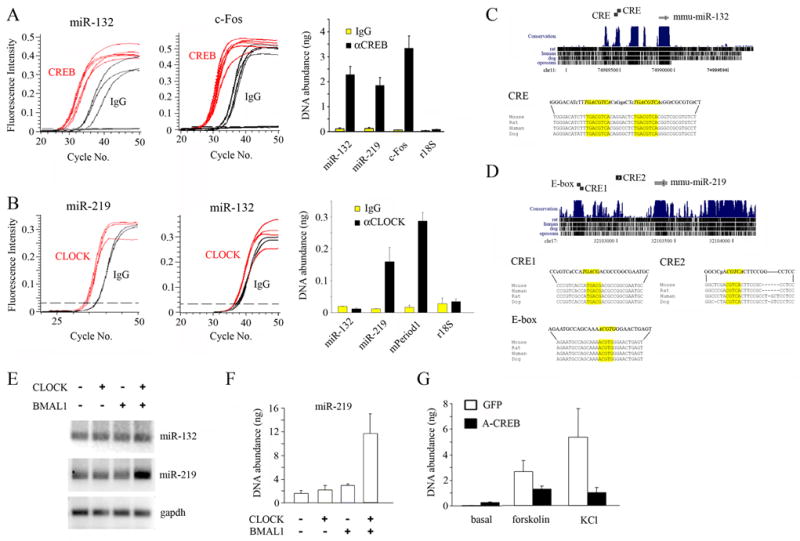
(A–B) Chromatin immunoprecipitation (ChIP) assay of PC12 cells using (A) an anti-CREB or (B) an anti-CLOCK antibody. RNA abundance was determined by real-time quantitative PCR. A nonspecific IgG was used as a negative control. In (A), the levels of immunoprecipitated (IP) DNA encompassing the promoter regions of miR-132, miR-219-1, c-Fos, and an r18S control were quantitated (right), and data for miR-132 (left) and c-Fos (middle) are presented as fluorescence intensity vs. number of PCR cycles. In (B), the levels of IP promoter DNA of miR-132, miR-219-1, mPeriod1 and an r18S control were determined (right), and data for miR-219-1 (left) and miR-132 (middle) are presented as fluorescence intensity vs. number of PCR cycles. (C) Position of murine (mmu-)miR-132 gene on chromosome 11 (top). Alignment scores are indicated by black bars in the conservation track of mouse, rat, human, dog and opossum (top). Two consensus CREs (italicized: yellow boxes) are located in the 5′ region of the miR-132 locus, and base mismatches from mouse in rat, human and dog are represented by lower case letters. Sequence alignments of mouse, rat, human and dog (bottom). (D) Position of murine (mmu-)miR-219-1 gene on chromosome 17 (top). Alignment scores are indicated in the conservation track (top). Two putative CREs (CRE1 and CRE2; italicized, yellow boxes) and an E-box motif (italicized, yellow box) are located in the 5′ region of the miR-219-1 locus (bottom). Mismatches from mouse are denoted by lower case letters. (E) Levels of pre-miR-219-1, but not of pre-miR-132 or gapdh, were elevated in PC12 cells co-transfected with expression constructs for CLOCK and BMAL1, as determined by semi-quantitative reverse transcription (RT)-PCR. (F) Quantitative analysis of CLOCK/BMAL1-dependent induction of pre-miR-219 in transfected PC12 cells as determined by real-time PCR. Data are presented as mean ± SEM nanograms of DNA. (G) Dominant-negative CREB (A-CREB) suppresses miR-132 induction. Primary cortical neurons were transfected with the expression constructs for A-CREB or GFP (negative control), were stimulated with forskolin (10 μM) or KCl (20 mM), and the abundance of pre-miR-132 transcript was determined by real-time PCR. Data are presented as mean ± SEM nanograms of cDNA.
