Figure 2.
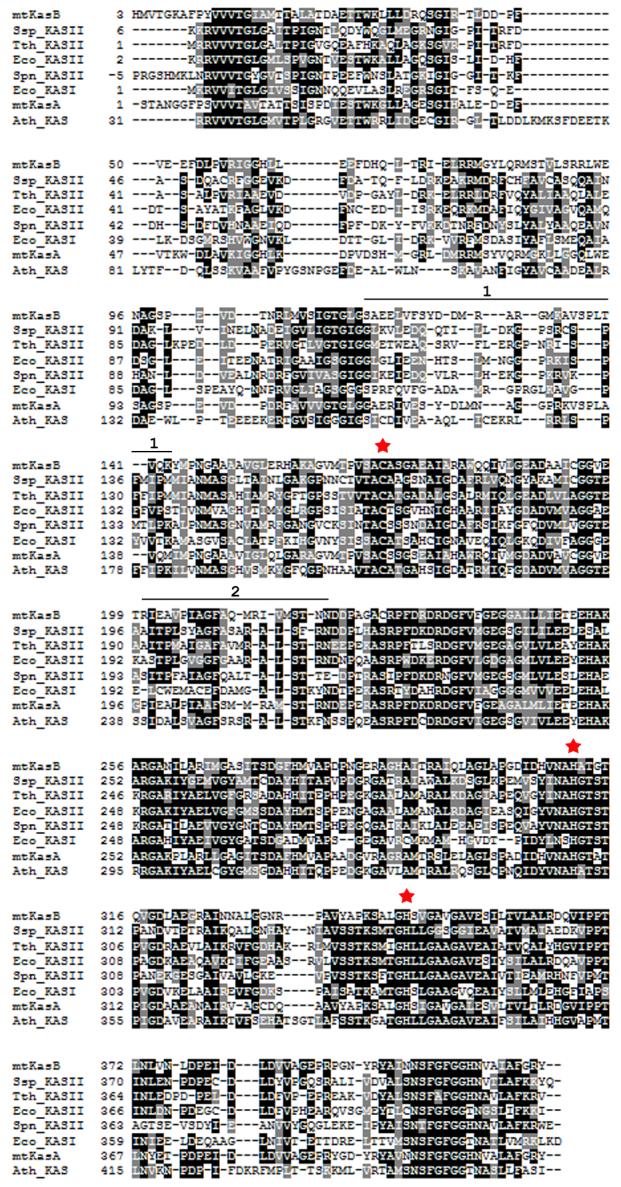
Structure based alignment of KAS-I/II sequences. A structure based alignment was generated for KAS-I/II sequences using Protein structure comparison service SSM at European Bioinformatics institute59 (http://www.ebi.ac.uk/msd-srv/ssm) and Boxshade (http://www.ch.embnet.org/software/BOX_form.html). The red stars mark the active site residues. The stretches of sequence marked by the black line above the mtKasB sequence and labeled 1 and 2 refer to the segments colored in black in Figure 1. Ssp_KASII - Synechocystis sp. KAS II (PDB code of the structure used in alignment: 1E5M, rmsd of alignment with mtKasB reported by the SSM service: 1.6 Å), Tth_KASII - T. thermophilus KAS II (1J3N, 1.5 Å), Eco_KASII - E.coli FabF (1KAS, 1.6 Å), Spn_KASII - S. pneumoniae KAS II (1OX0, 1.7 Å), Eco_KASI - E.coli FabB (1DD8, 1.7 Å), mtKasA - M. tuberculosis KasA (0.6 Å), Ath_KAS - A. thaliana mitochondrial KAS (1W0I, 1.7 Å).
