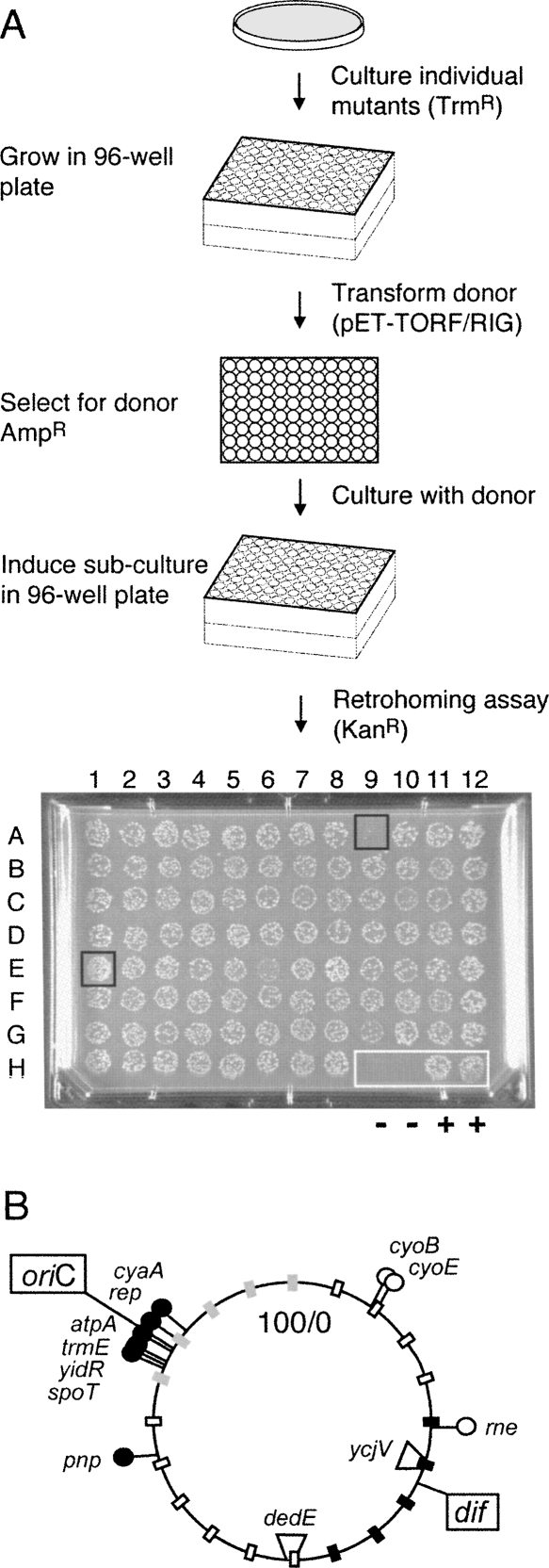
FIGURE 2.
Mutant host screen and location of Tn5 insertions. (A) Schematic of retrohoming assay to screen E. coli mutant libraries. Strain MC1061 dedE∷HS contains the −30/+15 homing site in the dedE gene. Individual trimethoprim-resistant MC1061 dedE∷HS colonies, containing a randomly integrated EZ∷TN <DHFR> transposon, were recovered in selective medium and stored in 96-well plates. Each mutant was then transformed with the pET-TORF/RIG-ampR donor plasmid, and fresh transformants were grown overnight. Ll.LtrB intron expression was induced for 3 h in diluted log-phase cultures, which were spotted onto omnitray slabs containing kanamycin and incubated overnight. A sample slab is shown with two negative (−) control spots at H9 and H10 and with two positive (+) wild-type control spots at H11 and H12. Spot E1 represents a candidate for an “up” mutant and A9 for a “down” mutant. (B) Location on the E. coli genome of Tn5 insertions that affect retrohoming in dedE∷HS and ycjV∷HS strains. The filled circles indicate mutations that decrease retrohoming, and the open circles are mutations that increase retrohoming. The circular map is marked as in Figure 1A, with the location of the homing-site genes used to determine retrohoming frequencies indicated (dedE and ycjV).