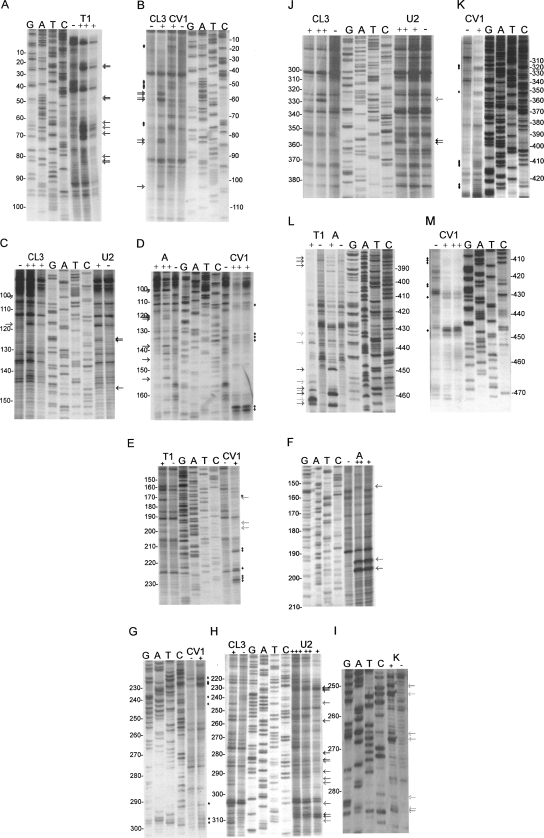
FIGURE 2.
Biochemical analysis of the FIV packaging signal region RNA. FIV Petaluma strain nt 1–511 were transcribed in vitro and were digested with RNases or chemicals specific for ss (A, T1, CL3, U2, kethoxal) or ds (CV1) nucleotides. Primers were annealed to the RNA and cDNAs were made with AMV RT, incorporating 33P-dATP. Samples were separated on 10% polyacrylamide gels, alongside cycle sequencing ladders. Sequence is given in the genome sense for clarity. Different primers were used to examine different regions of the RNA. These regions were: A, B: R/U5; C, D: SL1 and PBS; E, F, G, H, I: SL2; J, K: SL3, SL4, gag; L, M: gag. Numbers represent FIV nucleotide numbers shown in Figure 3. Various concentrations of each RNase/chemical or incubation time were used; only lanes where RNAs were minimally cleaved are shown. These were: A, RNase T1 0.2/1 U; B, RNase CL3 1 U and RNase CV1 2 min; C, RNase CL3 0.1/1 U, RNase U2 0.1 U; D RNase A 0.2/2 pg, RNase CV1 30 sec/2 min; E, RNase T1 0.1 U, CV1 30 sec; F, RNase A 0.2/2 pg; G, RNase CV1 5 sec; H, RNase CL3 1 U, RNase U2 0.01/0.1/ 0.5 U; I, kethoxal 200 μg; J, RNase CL3 0.1/0.2 U, RNase U2 0.1/0.2 U; K, RNase CV1 2 min; L, RNase T1 0.1 U, RNase A 0.2 pg; M, RNase CV1 30 sec/2 min. RNase cleavage sites are indicated with arrows (ss nucleases, colored light to dark gray in the order T1, CL3, A, U2) or diamonds (CV1).