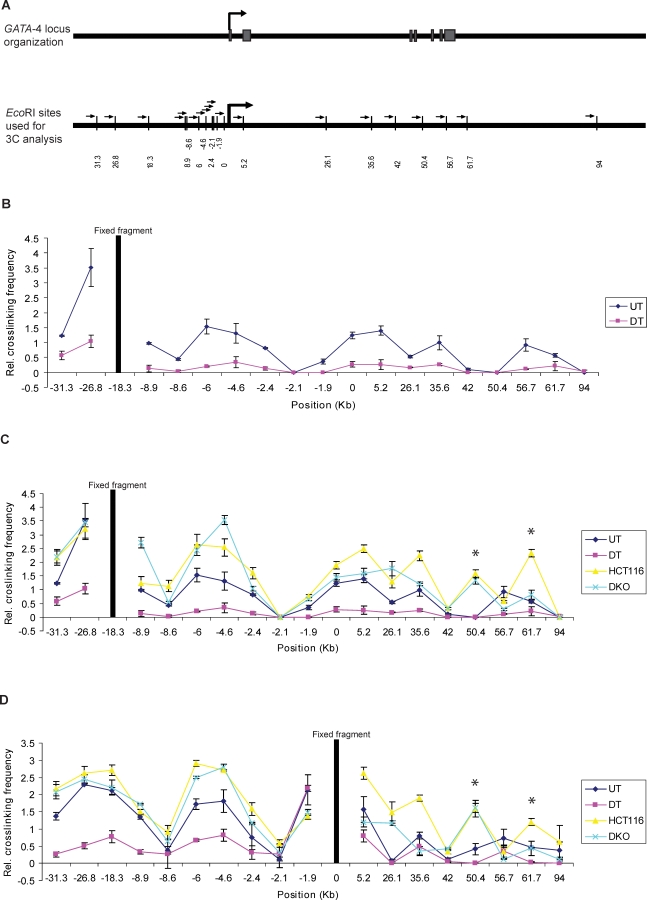
Figure 1. The Long-Range Chromatin Interactions at the GATA-4 Locus in Undifferentiated EC Cells, HCT116, and DKO Cells and Its Loss in Differentiated EC Cells, as Mapped by Chromosome Conformation Capture (3C) Assay.
(A) Representation of the genomic organization of the human GATA-4 locus (upper panel) and EcoRI sites used for this study (small vertical bars, lower panel), with distance in kb along the x-axis (lower panel). The seven exons of the gene are shown as grey boxes, intervened by six introns. The large arrowhead represents the transcription start site. The gene spans approximately 55 kbp on 8p23.1–22. The EcoRI site nearest to the transcription start site is 0 on the x-axis. The fragments at −18.3 (B and C) and promoter (0) (D) were used as bait in the 3C assay for bidirectional search of physical partners. The small arrows next to each EcoRI site represent the location and the direction of primers used for the 3C assay in Figures 1 and 4.
(B) The relative crosslinking frequency of different regions interacting with the −18.3 fragment (fixed fragment, the thick black line) across the GATA-4 locus in undifferentiated Tera-2 cells (blue dots) and is compared to EC cells differentiated by ATRA treatment (purple dots) . Standard error of the mean is indicated by the brackets around each dot, which represent the average derived from at least five independent samples. The value of relative crosslinking frequency is plotted on the y-axis and the locations of various EcoRI sites (in kb) used for the 3C analysis are plotted on the x-axis. The calculation of relative crosslinking frequency between two given GATA-4 fragments was done essentially as described by others [81], which allows a direct comparison between the different cell types used in the 3C assay by correcting for possible variants. ERCC3 fragments separated by approximately 10 kb were used for normalizing crosslinking frequency. UT= undifferentiated Tera-2 cells, DT= differentiated Tera-2 cells.
(C) The spatial organization of the GATA-4 locus in the four cell types examined: HCT116, DKO, undifferentiated Tera-2, and differentiated Tera-2 cells. The graph is plotted similarly to (B) (for explanation of symbols, refer to (B)). Relative crosslinking frequencies of different regions with the −18.3 fragment in undifferentiated Tera-2 cells (UT), differentiated Tera-2 (DT), HCT116, and DKO cells are shown as blue, purple, yellow, and green dots, respectively, as indicated to the far right of the panel. The two novel interactions discovered in HCT116 cells, located at +50.4 and +61.7 kb, are marked by asterisks, one of which (+61.7 Kb) had reduced crosslinking frequency in DKO cells.
(D) The results of 3C analysis in the four cell types using the promoter fragment (0) as bait, instead of the −18.3 fragment. The graph is plotted similarly to (B) (for explanation of symbols, refer to (B)). Relative crosslinking frequencies of different regions with the promoter (0) fragment in undifferentiated Tera-2 cells (UT), differentiated Tera-2 (DT), HCT116, and DKO cells are shown as blue, purple, yellow, and green dots, respectively, as indicated to the far right of the panel.