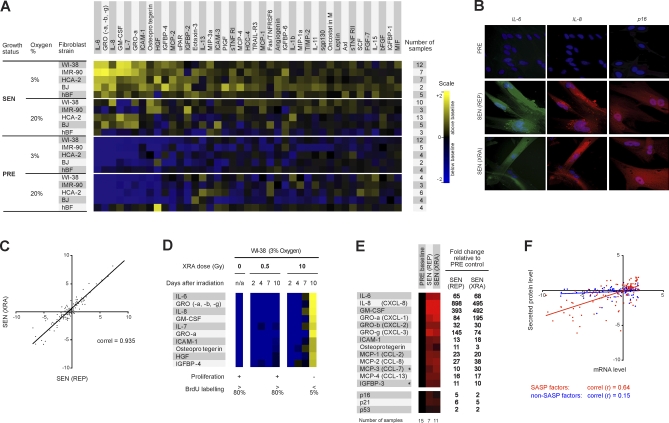
Figure 1. SASP of Human Fibroblasts.
(A) Soluble factors secreted by the indicated cells were detected by antibody arrays and analyzed as described in the text, Materials and Methods; Text S2; and Datasets S1–S4. For each cell strain, the PRE and SEN signals were averaged and used as the baseline. Senescence (SEN) was induced by either repeatedly passaging the cells (REP, replicative exhaustion) or by exposing them to a relatively high dose (10 Gy) of ionizing radiation (XRA, X-irradiation); for simplification, XRA and REP signals were averaged as a single SEN signal (see also Figure 1C). Signals higher than baseline are displayed in yellow; signals below baseline are displayed in blue. The numbers on the heat map key (right) indicates log2-fold changes from the baseline.
(B) Validation by immunostaining. PRE WI-38 cells were made senescent by REP or XRA, maintained in 10% serum until fixation, and immunostained for the cytokines IL-6 and IL-8 and the senescence marker p16INK4a.
(C) Correlation between the SASPs of WI-38 cells induced to senescence by XRA or REP, compared to PRE cells, depicted as log2-fold changes.
(D) WI-38 cells were X-irradiated at the indicated doses. CM was collected 2, 4, 7, or 10 d later, and soluble factors were analyzed by antibody arrays. PRE cells and cells irradiated with 0.5 Gy transiently arrested growth, but resumed proliferation 24–48 h after CM collection. Cells irradiated with 10 Gy became senescent, and therefore did proliferate during and beyond the course of the experiment. PRE and SEN (10 d) signals were averaged and used as the baseline. Signals higher than baseline are displayed in yellow; signals below baseline are displayed in blue.
(E) PRE and SEN cells were analyzed for the indicated mRNAs by quantitative RT-PCR, and normalized to the corresponding PRE values (baselines). The senescence inducers (REP or XRA) are given in parentheses. Signals higher than baselines are shown in red, signals below baselines are displayed in green, and the fold changes in signals are given to the right of the heat map. The results are averages for four cell strains (WI-38, IMR90, HCA-2, and BJ), and the total number of samples for each condition are given below the heat map. An asterisk (*) indicates non-SASP factors (see Figure S4).
(F) Correlation between mRNA and secreted protein levels for SASP (red) and non-SASP (blue) factors (see Figure S4). PRE and SEN values were averaged to create a baseline; all values were expressed as the log2-fold change relative to this baseline; PRE versus baseline and SEN versus baseline are shown.