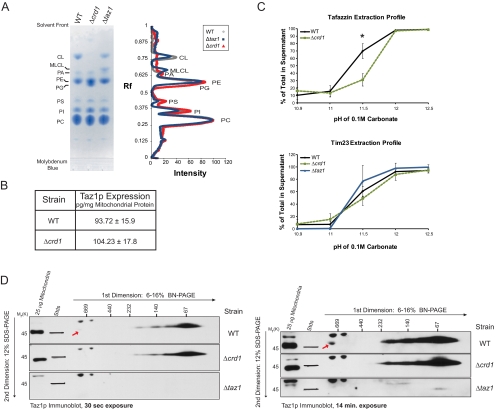
Figure 1.
Altered membrane association and complex formation of Taz1p in absence of CL. (A) Phospholipids were extracted from 1 mg of mitochondria isolated from the indicated strains and separated by TLC, and phospholipids were visualized using molybdenum blue. Scanned images were analyzed using Quantity 1 software and lane analyses functions. The determined phospholipid profile for each mitochondrial sample was calculated after background correction. The migration of phospholipids is indicated; PC, phosphatidylcholine; PI, phosphatidylinositol; PS, phosphatidylserine; PE, phosphatidylethanolamine; PA, phosphatidic acid. (B) Quantification of Taz1p levels in mitochondria was determined by immunoblot using a recHis6Taz standard curve (mean ± SEM, n = 4). (C) WT or Δcrd1 mitochondria were analyzed by alkali extraction using 0.1 M carbonate at the indicated pH values. Equal volumes of the pellet (P) and TCA-precipitated supernatant (S) fractions were immunoblotted for the indicated mitochondrial markers. Bio-Rad (Richmond, CA) Versadoc-captured images were quantified using the affiliated Quantity 1 software and the percent Taz1p or Tim23p present in the derived supernatants after extraction in carbonate at each pH determined as follows: S/(S+P) × 100, where S is the volume of protein detected in the supernatant at a given pH and P is the volume associated with the pellet at the same pH. The asterisks indicates a statistically significant difference in the extractability of Taz1p from wt and Δcrd1 mitochondria as calculated by the Student's t test, p < 0.001 (mean ± SD, n = 4–5). (D) 100 μg of 1.5% (wt/vol) digitonin extracts from mitochondria derived from the indicated strains were resolved by 2D BN/SDS-PAGE, and Taz1p complexes were revealed by immunoblot. The exposure of each set of immunoblots is designated at the bottom. n = 3.