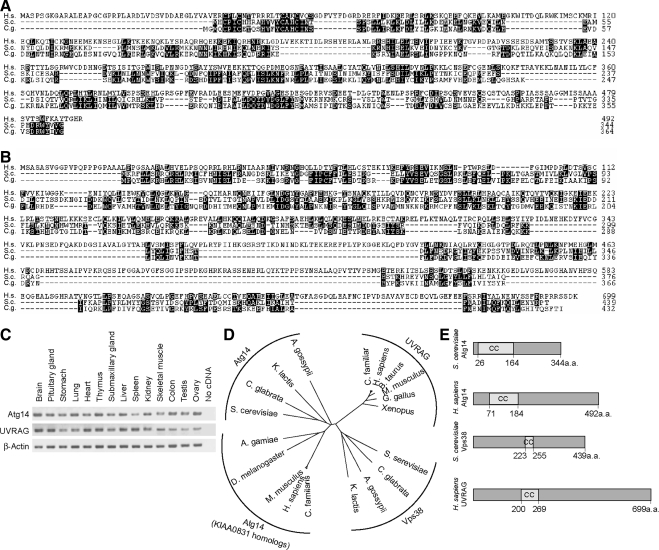
Figure 1.
Identification of mammalian counterparts of Atg14 and Vps38. (A) Amino acid alignment of Homo sapiens Atg14 (KIAA0831; National Center for Biotechnology Information [NCBI]accession no. NP_055739) with Atg14 of S. cerevisiae (accession no. NP_009686) and C. glabrata (accession no. XP_445209). The alignment was generated using CLUSTAL W. Identical residues are indicated with filled boxes. (B) Amino acid alignment of H. sapiens UVRAG (NCBI accession no. NP_003360) with Vps38 of S. cerevisiae (accession no. NP_013464) and C. glabrata (accession no. XP_445209). (C) KIAA0831 (Atg14) and UVRAG are ubiquitously expressed in mouse tissues. Total RNA from mouse tissues was reverse-transcribed into cDNA and then subjected to PCR amplification with indicated primers. (D) Phylogenetic analysis of Atg14, KIAA0831, UVRAG, and Vps38 homologues. The unrooted phylogenetic tree was constructed using CLUSTAL W based on the amino acid sequences of these homologues. Distance matrix-based trees were constructed by the NJ method (Saitou and Nei, 1987). (E) Structural comparison of S. cerevisiae Atg14, H. sapiens Atg14 (KIAA0831), S. cerevisiae Vps38, and H. sapiens UVRAG. The coiled-coil domain is predicted by the algorithm of Lupas et al. (1991).