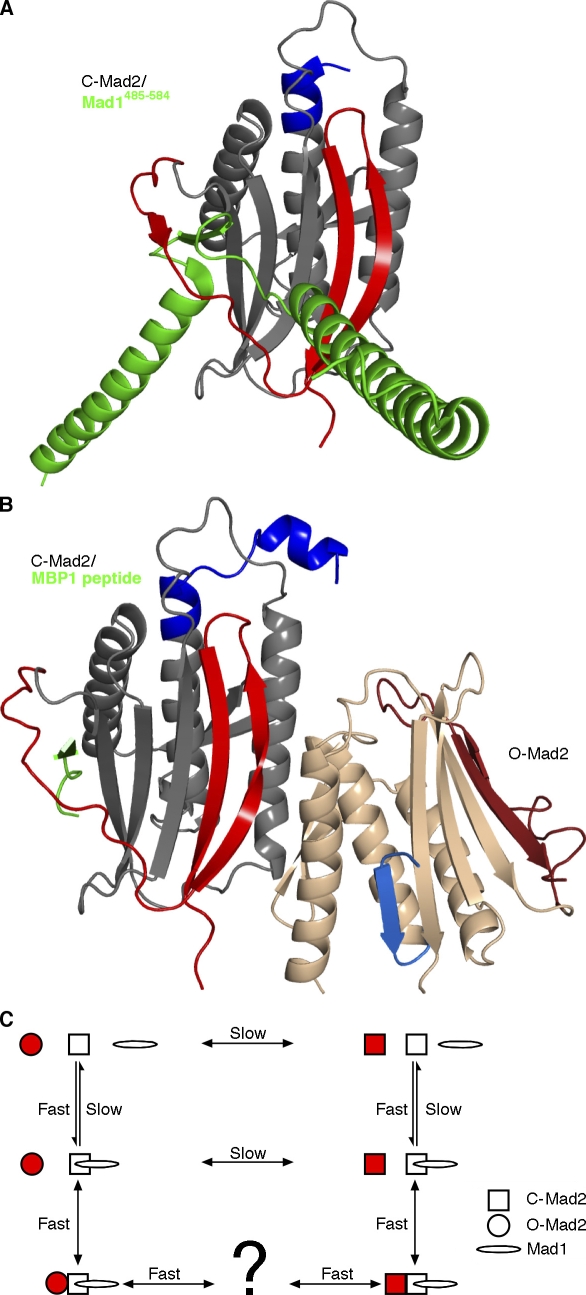
Figure 2.
Mad2-containing complexes. (A and B) The displacement of the C-terminal segments in C-Mad2 exposes a new β sheet edge that can incorporate Mad1 (A; PDB 1GO4; Sironi et al., 2002), Cdc20, or the synthetic peptide MBP1 (B; PDB 2V64; Mapelli et al., 2007) between newly exposed β6 and newly formed β7′. In the crystal structure of the O-Mad2–C-Mad2 dimer (B), asymmetrical dimerization occurs mainly through the unaltered core of Mad2 (gray to tan) but also includes the β8′ strand that is unique to C-Mad2 (coloring as in Fig. 1 C). O-Mad2, the form undergoing conversion (tan), interacts with C-Mad2 only through its unchanging core. (C) A reaction scheme for Mad2 catalysis. Mad2 (red) represents the molecule undergoing conversion. Uncatalyzed Mad2 interconversion proceeds far more slowly (lifetime >9 h; Luo et al., 2004) than the duration of metaphase (∼20 min). The Mad2 structural rearrangement is catalyzed by binding to the Mad1–C-Mad2 complex. In this reaction scheme, catalysis by induced fit would increase the forward O-Mad2→C-Mad2 rate, whereas the conformational selection of C-Mad2 would reduce the reverse O-Mad2←C-Mad2 rate. It is unknown whether Mad2 releases from the Mad1–C-Mad2 dimer as fully folded C-Mad2 or as a partially unfolded intermediate.