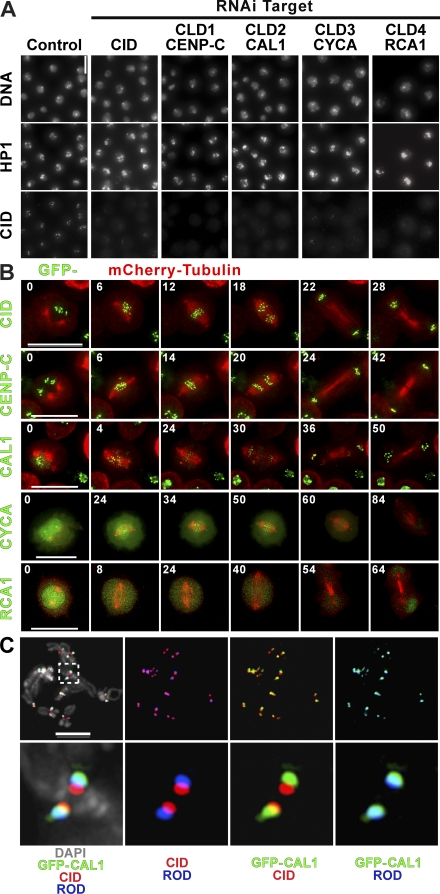
Figure 1.
Identification and localization of CLDs. (A) IF of cells depleted of the top four positive hits from the screen. Cells were stained for DNA, CID, and HP1. CID localization to the centromere (bottom) is highly reduced or absent after RNAi depletion of all four candidates or CID in comparison with the control (left), and HP1 staining is normal in all cases. Notice that in addition to CID depletion, RCA1 and CYCA RNAi result in endoreduplication and an increase in nuclear size. (B) CLD dynamics through mitosis. Still images from time-lapse analysis of stable S2 cell lines expressing mCherry-tubulin (red) and GFP-CID, –CENP-C, -CAL1, -CYCA, and -RCA1; relative times in minutes from the start of the video are indicated in each frame. See Videos 1–5 (available at http://www.jcb.org/cgi/content/full/jcb.200806038/DC1). (C) Localization of GFP-CAL1, CID, and Rod. Metaphase chromosome spreads of S2 cells stably expressing GFP-CAL1 were stained with anti-CID (red), anti-GFP (green), and anti-Rod (blue) antibodies. The GFP-CAL1 signal overlapped significantly with the outer kinetochore protein Rod, whereas only a little overlap was observed with CID, indicating that CAL1 is located close to the outer kinetochore in mitotic chromosomes, which is in contrast to localization to CID chromatin in interphase. Inset shows magnification of image. Bars: (A) 15 μm; (B and C) 5 μm.