Abstract
Cultivation-independent analyses based on genetic profiling of partial bacterial 16S rRNA genes by PCR-single-strand conformation polymorphism (PCR-SSCP), reverse transcriptase (RT)-PCR-SSCP of the 16S rRNA itself, and stable isotope probing (SIP), followed by RT-PCR-SSCP, were applied to characterize the diversity of metabolically active bacteria in the larval gut of Manduca sexta bred on tobacco leaves under greenhouse conditions. For SIP, hatching larvae were fed with leaves from tobacco plants grown in a 13CO2-enriched atmosphere. Dominant SSCP bands were sequenced and phylogenetically analyzed. Only one major gut colonizer, an Enterococcus relative, was detected; it occurred in the heavy RNA fraction, demonstrating its metabolic activity, and it originated from eggs, where its metabolic activity was also indicated by rRNA-based SSCP profiles. In contrast, a Citrobacter sedlakii relative was detected on eggs by DNA-SSCP, but rRNA-SSCP and SIP-rRNA-SSCP were negative, suggesting that these bacterial cells were inactive. A Burkholderia relative was dominant and metabolically active on the tobacco leaves but inactive inside the gut, where it was also quantitatively reduced, as suggested by lower band intensities in the DNA-based SSCP profiles. SIP-RNA-SSCP detected another metabolically active gut bacterium (Enterobacter sp.) and more bacteria in the light RNA fraction, indicating low or no metabolic activity of the latter inside the gut. We conclude that the larval gut supported only a low diversity of metabolically active bacteria.
Insects represent one of the largest reservoirs of bacterial diversity on Earth, and more than 250 million years of coevolution have resulted in manifold interactions between insects and bacteria, ranging from pathogenicity to highly sophisticated symbiotic relationships (13, 56). One of the most striking interactions is that bacteria have extended the nutritional range of insects by supplying nutrients as endosymbionts (14) and by accessing otherwise indigestible substrates, such as lingo-cellulose-derived organic matter from soils, with the help of gut bacteria (7).
Manduca sexta (Sphingidae) represents a lepidopteran species adapted to extreme habitat conditions because their larvae can feed exclusively on the leaves of tobacco plants (43). Tobacco plants typically contain high amounts of alkaloids which are highly toxic to other insects and vertebrates but not to M. sexta (21, 49, 57). Once hatched from eggs deposited on tobacco leaves, the M. sexta larvae consume large amounts of such leaves and pass through five larval stages (instars) within 2 to 3 weeks, increasing their individual body weight from approximately 1 mg to 10 g (22, 43). While this simple feeding strategy is a large problem for tobacco growers (23), it is, under controlled conditions, an excellent system to generate larvae for biological research, which explains the broad use of M. sexta in laboratories worldwide (for examples, see references 26, 29, and 52).
Despite their wide laboratory dissemination and their economic importance as plant pests, surprisingly little is known about the bacterial communities in their guts. Cultivation studies suggest that the diversity is low, as only two 16S rRNA types were retrieved on LB plates, but these were not characterized further, e.g., by DNA sequencing (10). In another study, three different bacterial strains, in addition to a yeast strain, were cultivated from M. sexta larval gut contents after incubation in a nutrient broth and on LB agar. These strains were tentatively assigned to two Bacillus species and a Serratia plymuthica (54). A survey of gut and fecal bacteria culturable under aerobic conditions on common laboratory media and characterized by 16S rRNA gene sequencing retrieved relatives of the genera Enterobacter, Escherichia, Pantoea, Flavobacterium, Pseudomonas, and Bacillus (unpublished results; accession numbers in the EMBL nucleotide database, FM212207 to FM212221). Experience from many similar studies suggests that such cultivation data most likely give a wrong picture of the actual microbial diversity, which makes it rewarding to use methods independent of cultivation.
The gut of M. sexta, just like that of other herbivorous lepidopteran larvae (caterpillars), is a simple tube comprising the largest part of the whole body. This simple structure can be explained by phylogenetic adaptation for the rapid passage and processing of large quantities of solid plant material (15). Midgut conditions are highly alkaline with pH values of 9 and above, which facilitates the extraction of nutrients (2). It can be assumed that such alkaline conditions along with frequent quick molting cycles between larval stages, which also affect gut function (8), and speedy food passage altogether present relatively selective conditions for bacterial communities. A recent cultivation-independent analysis of another lepidopteran larva (Lymantria dispar) in fact revealed the presence of relatively low diversity with only nine different 16S rRNA sequences (phylotypes) (6).
The objective of this study was to characterize the bacterial diversity in the guts of M. sexta larvae kept under laboratory breeding conditions. The study was based on the cultivation-independent detection of PCR-amplified partial 16S rRNA sequences and their phylogenetic assignment to look for closest relatives. In order to determine the source of the bacteria, the bacterial diversity associated with the food (tobacco leaves) and with M. sexta eggs deposited on the leaf surfaces were also determined. To allow the distinction between abundant and metabolically active bacteria, comparisons were made between genetic profiles with the single-strand conformation polymorphism (SSCP) technique (12, 48) of the partial 16S rRNA genes PCR amplified from the total community DNA and 16S rRNA itself using reverse transcriptase PCR (RT-PCR). In addition, it was explored whether stable isotope probing (SIP) of 16S rRNA sequences (34) would also be capable of and possibly more sensitive in detecting metabolically active gut bacteria. For this purpose, larvae were bred on the leaf material of tobacco plants previously grown in an atmosphere highly enriched with 13CO2.
MATERIALS AND METHODS
Identity and cultivation of the tobacco plants.
All feeding experiments in this study were conducted with Nicotiana tabacum cv. PBD6 (nontransgenic) plants and their corresponding plastid transformants (transgenic), carrying aadA genes. Details of the plants and their construction have been described elsewhere (30). The plants, if not otherwise stated, were grown from seeds in pots with a diameter of 30 cm filled with potting soil (Einheitserde Typ ED 73, Einheitserdewerke, Hameln, Germany) at 28°C under a light-dark cycle of 12 h each per day. During cultivation, all plants were fertilized with 0.2% (vol/vol) Manna Wuxal Super (Haug, Ammerbuch-Pfäffigen, Germany). Seeds of nontransgenic and transgenic plants were kindly provided by the TRANSBAC coordinators at the Institute Écologie Microbienne, Université Claude Bernard Lyon, France.
13C-enriched tobacco plants were generated under greenhouse conditions to supply 13CO2 as a plant carbon source. For this purpose, seeds of nontransgenic and transgenic N. tabacum were surface sterilized by rinsing with a 70% (vol/vol) aqueous ethanol solution for 2 min, followed by an incubation of 5 min in 12% (vol/vol) sodium hypochlorite, before washing five times with sterile distilled water. The seeds, kept under sterile conditions, were germinated at 26°C for a photoperiod of 16 h up to the first leaf stage, before seedlings were transferred into 50-ml Falcon tubes (Becton Dickenson, Heidelberg, Germany) half filled with sterile expanded clay (grain size, 4 to 8 mm). Each tube containing a seedling was closed with Parafilm (Brand, Wertheim, Germany), and plants were cultivated for an additional 2 weeks. Sterile water and liquid fertilizer (0.2% [vol/vol] Manna Wuxal Super) were added to each plant as recommended.
13CO2 was produced by weighing 1.4 g “heavy” sodium carbonate (Na213CO3; Sigma-Aldrich Chemie, Taufkirchen, Germany) and 500 μl of 3% (wt/vol) methyl orange (Merck, Darmstadt, Germany) into a bottle with a volume of 316 ml. The bottle was evacuated, and 7 ml of 10% (vol/vol) sulfuric acid (H2SO4) was injected through a membrane inserted in the lid of the bottle to generate 13CO2. The amounts of sodium carbonate and sulfuric acid were calculated to fill the bottle completely with 13CO2. To avoid a negative pressure in the bottle, the volume of the removed gas was replaced by the addition of 10% (vol/vol) H2SO4.
Four-week-old tobacco plants, six transplastomic and six isogenic control plants were transferred into a gas-tight chamber. The chamber was connected via a Tygon plastic tube to an infrared carbon dioxide analyzer (LCA2, ADC BioScientific, Great Amwall, United Kingdom), measuring the actual CO2 concentration in this closed system. The chamber was set up in the greenhouse at 26°C; depending on the external lighting conditions, artificial light was added as required. The circulation of air was achieved by a ventilator attached to the top of the chamber; the temperature inside was controlled using a thermometer. 13CO2 was injected with a syringe into the chamber up to a maximum carbon dioxide concentration of 1,000 mg liter−1, and when a minimum of 200 mg liter−1 was reached, more 13CO2 was added. Twelve plants were grown under these conditions for 12 h each day over a period of 15 days. The plants were kept in a closed box in the dark for 12 h each night and thereby protected from photosynthetically active light. Water and liquid fertilizer were added as required.
Feeding experiments.
Breeding stocks of Manduca sexta were kept as described previously (5). Eggs from these breeding stocks were directly transferred onto whole tobacco plants. After hatching, the larvae began to feed on the plant material and went through all five larval stages within the following 5 weeks. Five to 10 larvae of each stage were collected and analyzed. Larvae of the instar 1 and instar 2 stages were used as a whole for the extraction of total nucleic acids, while for the following three larval stages, total nucleic acids were extracted from the guts only. Depending on the larval stage, guts or whole larvae were stored in 70% ethanol at 4°C for 3 days prior to the extraction of the total nucleic acids. In addition, 100 eggs of M. sexta were conserved in 70% ethanol and stored at −20°C. Also, feces from each larval stadium were collected and stored at 4°C for up to 3 days before extraction of the total nucleic acids.
Total nucleic acid extractions for bacterial community analyses.
Gut and feces samples, as well as eggs and tobacco leaves, were ground in liquid nitrogen using a mortar and pestle. The crushed material was stored at −70°C until further processing. The extraction of total nucleic acid was conducted by using the protocol of Lueders et al. (33) with 100 mg of each ground sample. The quality of the total nucleic acids was evaluated on 0.8% (wt/vol) agarose gel (44). Nucleic acid extracts were stored in aliquots at −20°C.
PCR amplification of the partial 16S rRNA gene sequences.
To generate genetic profiles with SSCP (see below), partial 16S rRNA gene sequences were amplified by PCR from total nucleic acids using a nested-PCR approach. The first PCR was conducted with primers 27f and 1492r (55) to amplify the almost complete bacterial 16S rRNA gene. The second PCR was used to generate the PCR products for the SSCP profiles with the universal Com1 and Com2-Ph primers, the latter primer being phosphorylated for further single-strand digestion (48). This nested PCR approach was chosen to exclude the amplification of 18S rRNA genes from contaminating gut tissue. Each PCR was conducted in a total volume of 50 μl. The PCR master solution contained 0.5 μM concentrations of each primer (MWG Biotech, Ebersberg, Germany), each nucleotide at a concentration of 0.2 mM (Qbiogene, Heidelberg, Germany), and 1.25 U of Taq polymerase (HotStar Taq; Qiagen, Hilden, Germany) with the corresponding 1× PCR buffer, 1.5 mM MgCl2, and 1.5 μl template DNA. PCR amplifications were conducted in a Primus96 thermocycler (MWG Biotech). An initial denaturation step at 95°C for 15 min was followed by 30 cycles of 60 s at 94°C, 60 s at 50°C, 70 s at 72°C, and, for the last cycle, a final extension of 5 min at 72°C was added. The second PCR with primers Com1 and Com2-Ph was conducted with only 25 cycles under the same conditions. For each sample, PCR amplifications were carried out in triplicate, and products were analyzed by agarose gel electrophoresis stained with ethidium bromide (44) and then combined.
Amplification of the 16S rRNA sequences by RT-PCR.
For rRNA-based genetic profiles, DNA was digested from the total nucleic acid samples using the RQ1 RNase-free DNase (Promega, Mannheim, Germany), which degrades both double- and single-stranded DNA endonucleolytically, producing 3′-OH oligonucleotides. Before digestion, DNA concentrations in the total nucleic acid samples were determined with the PicoGreen double-stranded DNA quantification reagent (MoBiTec, Göttingen, Germany) and a microtiter plate reader (Fluroskan II; Labsystems, Helsinki, Finland). The concentration of RQ1 RNase was adjusted to the DNA concentration as recommended by the manufacturer. For each sample, a DNase digestion reaction mix was set up by using up to 8 μl of each sample (total nucleic acid), 1 μl RQ1 RNase-free DNase 10× reaction buffer, and 1 U μg−1 DNA of the RQ1 RNase-free DNase. Each digestion approach was incubated at 37°C for 30 min using a Primus96 thermocycler (MWG Biotech). After incubation, 1 μl of RQ1 DNase Stop solution per reaction terminated the digestion reaction, and a following incubation at 65°C for 10 min inactivated the DNase. RNA was stored at −70°C until further processing for a maximum of 3 days. After DNA digestion, the RNA samples were used as templates for a control PCR with primers 27f and 1492r followed by Com1 and Com2-Ph, respectively, as described above, to control the DNA digestion. Successful DNA digestion was indicated by the lack of a PCR product on the agarose gels.
The RNA concentration of each sample was determined by quantification using the RiboGreen quantification kit (MoBiTec) following the manufacturer's instructions. RT-PCR was performed by using the Access RT-PCR system (Promega). Each PCR was conducted in a total volume of 50 μl. The PCR master solution contained 1 μM of each primer (27f/1492r), each nucleotide at a concentration of 0.2 mM, 1 mM MgSO4, 0.1 U μl−1 avian myeloblastosis virus (AMV) RT, 0.1 U μl−1 Tfl polymerase, and the corresponding 1× AMV/Tfl reaction buffer. The reaction was initiated by adding 2 μl of RNA sample (10 pg to 1 μg). The first-strand cDNA synthesis was conducted by reverse transcription during incubation at 45°C for 45 min, followed by one cycle at 94°C for 2 min to allow AMV RT inactivation and RNA/cDNA/primer denaturation. The generation of the second-strand and PCR amplifications was conducted by 40 cycles of 30 s at 94°C, 1 min at 60°C, and 2 min at 68°C and a final extension of 7 min at 68°C. For a positive control, the supplied positive-control RNA with carrier and the upstream and downstream control primers were used. For a negative control, sterile nuclease-free water was substituted instead of RNA template. All steps were conducted in a Primus96 thermocycler (MWG Biotech). The PCR products from this reaction were used as a template for the following PCR (nested approach) using Com1 and Com2-Ph. PCR amplifications were carried out in triplicate, and products were analyzed by gel electrophoresis and combined.
Isopycnic centrifugation and fractionation of density gradients.
12C- and 13C-labeled RNA were separated by density gradient centrifugation using cesium trifluoroacetate (CsTFA) gradients (33). Density gradient centrifugation was performed in 4.9-ml OptiSeal polyallomer centrifuge tubes (Beckman Coulter, Krefeld, Germany) in a VTi 65.2 rotor (Beckman Coulter) in an ultracentrifuge (L8-70 M; Beckman Coulter). Centrifugation was conducted at 20°C and approximately 70 h at 39,000 rpm to form the CsTFA density gradients. Centrifugation mixtures were prepared by mixing 4.5 ml of a 2-g ml−1 CsTFA stock solution (Amersham Pharmacia Biotech, GE Healthcare Europe GmbH, München, Germany), 175 μl formamide, up to 1 ml gradient buffer (0.1 M Tris-HCl [pH 8], 0.1 M KCl, 1 mM EDTA), and RNA (up to 900 ng) to a final volume of 5.675 ml (the rRNA volume was subtracted from the volume of the gradient buffer). The density of the prepared mixtures was controlled with an AR200 digital refractometer (Leica Microsystems, Bensheim, Germany). The mixtures were transferred with a 10-ml syringe and a needle to the centrifuge tubes before the tubes were balanced and closed.
After centrifugation, the gradients were fractionated from the bottom to the top into 14 fractions of 375 μl each. For the collection of the gradient fractions, the centrifuge tubes were pierced from below with a sterile needle, and the removed gradient volume was replaced by water added from the top of the tube. The density of each fraction was determined by measuring aliquots of 75 μl in the refractometer. RNA was precipitated from all fractions with 500 μl isopropanol, followed by centrifugation for 30 min at 14,000 rpm at room temperature (Sigma 4K10, Osterode, Germany). The precipitates were washed with 150 μl ethanol (70%, vol/vol), spun for 5 min at 14,000 rpm, and resuspended in 30 μl of Tris-EDTA buffer. Reverse transcription of the RNA into cDNA and PCR amplification were performed as described above. The following second PCR to amplify the partial rRNA region used for the SSCP profiles was also conducted as described.
SSCP analysis.
Purification of the PCR and RT-PCR products was done with the QIAquick PCR purification kit (Qiagen) following the manufacturer's instructions. Each product was eluted in 30 μl elution buffer. After purification, DNA concentrations were determined with PicoGreen as described above. Approximately 800 ng of each purified PCR product was then treated at 37°C for 60 min with 5 U of lambda-exonuclease (Amersham Biosciences, GE Healthcare Europe GmbH, München, Germany) to remove the phosphorylated reverse DNA strand from the double-stranded PCR products (12). The single-stranded DNA molecules were purified using the MiniElute PCR purification kit (Qiagen) and eluted with 10 μl elution buffer before adding 8 μl of loading buffer (95% [vol/vol] formamide, 10 mM NaOH, 0.25% [wt/vol] bromophenol blue, and 0.25% [wt/vol] xylene cyanole). The samples were denatured for 2 min at 95°C, cooled on ice, and loaded on a nondenaturing polyacrylamide gel for SSCP electrophoresis. The pouring and staining of the SSCP gels as well as the electrophoretic conditions have been described elsewhere (47, 48). Briefly, the single-strand PCR products were separated in polyacrylamide gels (MDE; FMS Bioproducts) using a Macrophor apparatus (Amersham Biosciences) set at 400 V and 8 mA and a running temperature of 20°C for 6,800 V h. DNA was visualized by silver staining. SSCP markers were generated from bacterial pure cultures by PCR using Com1 and Com2-Ph and subsequent single-strand removal (see above).
Extraction and sequencing of bands from SSCP profiles.
Bands of interest were cut out from the silver-stained polyacrylamide SSCP gels with a sterile scalpel. The single-stranded DNA was eluted from the gel by the “crush and soak” procedure (44), resuspended in 12 μl of 10 mM Tris-HCl (pH 8.0), and amplified by PCR using primers Com1 and Com2-Ph under the conditions described above. The PCR products were purified using the NucleoSpin extract kit (Macherey-Nagel, Düren, Germany). After purification, ligation of the PCR products with the pGEM-T vector system (Promega) followed, and the ligated plasmids were transformed into Escherichia coli JM 109 for cloning, using the procedure recommended by the manufacturer (Promega). DNA of the clones with the vector inserts was extracted by alkaline lysis (15 min at 95°C in 0.05 M NaOH, 0.025% [wt/vol] sodium dodecyl sulfate [Sigma Aldrich]) and treated as a template in the PCR with primers puc18f (5′-CAC GAC GTT GTA AAA CGA C-3′) and puc18r (5′-GGA TAA CAA TTT CAC ACA GG-3′), hybridizing to the insert-flanking regions of the vector. A nested PCR of the diluted PCR obtained with primers Com1 and Com2-Ph followed. These PCR products were run on SSCP gels to compare the band position in the original community profile. Clones which carried the expected inserts were cultivated overnight in LB broth (44) containing ampicillin (final concentration, 100 mg liter−1), and plasmids were extracted with a plasmid DNA purification kit (Macherey-Nagel), following the protocol of the manufacturer. The sequencing reactions were carried out with the Sequitherm Excel II DNA sequencing kit-LC (Epicenter Technologies), and sequences were run and read using a LI-COR DNA 4200 GeneRead IR apparatus (LI-COR Biosciences, Bad Homburg, Germany) as described elsewhere (46).
Phylogenetic analyses.
Sequences were analyzed and aligned with the ARB software package (www.arb-home.de) (32). Sequences were compared to each other and to database sequences using BLASTN search analyses (http://www.ncbi.nlm.nih.gov/blast/) and tools provided by the Ribosomal Database Project RDP II (http://rdp.cme.msu.edu/).
Nucleotide sequence accession numbers.
All sequences of this study have been submitted to the Nucleotide Sequence Database of the European Bioinformatics Institute (www.ebi.ac.uk). The accession numbers are listed in Table 1.
TABLE 1.
Characterization of DNA sequences detected in this study after PCR or RT-PCR amplification from total nucleic acids extracted from feed (tobacco leaves), eggs, larval guts, or feces of Manduca sexta
| Band | Name of cloned sequence | Accession no. | Length of sequence (bp) | Closest relative(s) (% sequence identity) | Order (phylum or class) | Comment |
|---|---|---|---|---|---|---|
| D-SSCPa | ||||||
| 1 | Ms01aDNA | AM910351 | 369 | Burkholderia sp., Ralstonia sp., Cupriavidus sp. (all 99.7) | Burkholderiales (Betaproteobacteria) | Also in RNA profiles (band 5) |
| 2 | Ms08cDNA | AM910352 | 371 | Enterococcus gallinarum, Enterococcus casseliflavus, Enterococcus saccharolyticus (all 100) | Lactobacillales (Firmicutes) | Also in RNA profiles (band 6) |
| 3 | Ms09aDNA | AM910353 | 370 | Citrobacter sedlakii (99.7) | Enterobacteriales (Gammaproteobacteria) | Also in RNA profiles (band 9) |
| 4 | Ms10aDNA | AM910354 | 367 | Various uncultured clones (100) | Sphingobacteriales (Bacteroidetes) | Only in DNA profiles |
| R-SSCPb | ||||||
| 5 | Ms18cRNA | AM910355 | 369 | Burkholderia sp., Ralstonia sp., Cupriavidus sp. (all 98.6) | Burkholderiales (Betaproteobacteria) | 98.4% identical to band 1 |
| 6 | Ms21aRNA | AM910356 | 371 | E. gallinarum, E. casseliflavus, E. saccharolyticus (all 99.7) | Lactobacillales (Firmicutes) | 99.7% identical to band 2 |
| 7 | Ms22aRNA | AM910357 | 369 | Caulobacter sp. (100) | Caulobacterales (Alphaproteobacteria) | Weak band, only in RNA profiles |
| 8 | Ms01cRNA | AM910358 | 370 | Various Pseudomonas spp. (99.2) | Pseudomonadales (Gammaproteobacteria) | Weak band, only in RNA profiles |
| 9 | Ms28cRNA | AM910359 | 370 | C. sedlakii (100) | Enterobacteriales (Gammaproteobacteria) | 100% identical to band 3 |
| S-R-SSCPc | ||||||
| 10 | Ms10bSIP | AM910360 | 370 | Enterobacter cloacae, Enterobacter aphidicola, Enterobacter aerogenes (all 100) | Enterobacteriales (Gammaproteobacteria) | Only SIP-SSCP profiles (heavy fraction, indicating activity) |
| 11 | Ms16cSIP | AM910361 | 371 | E. gallinarum, E. casseliflavus, E. saccharolyticus (all 99.7) | Lactobacillales (Firmicutes) | 100% identical to band 2 |
| 12 | Ms15aSIP | AM910362 | 370 | Uncultured Sphingomonas sp. (98.9) | Sphingomonadales (Alphaproteobacteria) | Only in SIP-SSCP profiles; however, not in fractions, indicating activity |
| 13 | Ms17cSIP | AM910365 | 365 | Flavobacterium hydatis, other Flavobacterium spp. (100) | Flavobacteriales (Bacteroidetes) | |
| 14 | Ms03bSIP | AM910363 | 369 | Delftia acidovorans (100) | Burkholderiales (Betaproteobacteria) | |
| 15 | Ms11cSIP | AM910364 | 371 | Bacillus licheniformis (100) | Bacillales (Firmicutes) |
D-SSCP, isolated from DNA-based PCR SSCP profiles.
R-SSCP, isolated from RNA-based RT-PCR SSCP profiles.
S-R-SSCP, isolated from SIP RNA-based RT-PCR SSCP profiles.
RESULTS
SSCP profiles of PCR-amplified partial 16S rRNA genes from total DNA.
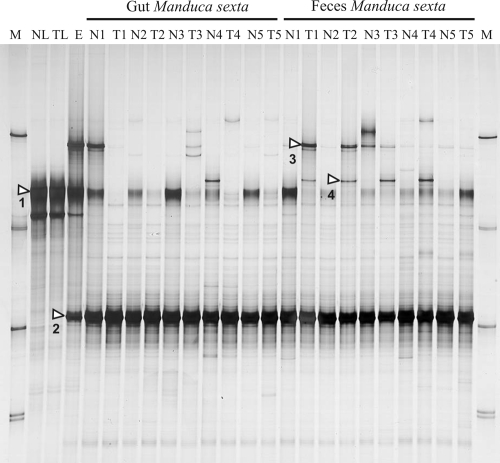
SSCP profiles were generated from the total DNA extracted from the tobacco leaves, larval eggs attached to the leaf surfaces, larvae or larval guts, and larval feces, respectively. The SSCP profiles of the tobacco leaves, the only food source supplied to the larvae, revealed the dominance of only one band (Fig. 1, band 1) which was characterized after cloning and DNA sequencing as originating from a member of the order Burkholderiales with the genera Burkholderia and Cupriavidus (previously Ralstonia) as the closest relatives (Table 1). Another less-intense band, below the dominant band, originated from the mitochondrial 16S rRNA genes of Nicotiana tabacum (data not shown). The SSCP profiles of eggs also contained both bands and, in addition, two other bands, one in a position close to the upper SSCP marker and the other running slightly above the third (Fig. 1, band 2). Band 2 was also the most-dominant band in all the SSCP profiles from the guts and feces. An SSCP band running in the same position as the upper band in the SSCP profiles from the eggs (Fig. 1, band 3) was also seen in the profiles from the guts and feces but only infrequently and in various intensities, independent of whether the leaves were transgenic or not. Sequencing and phylogenetic analysis revealed that the partial rRNA sequence of this band was closely related to the 16S rRNA gene of Citrobacter sedlakii (Table 1).
FIG. 1.
SSCP profiles of partial 16S rRNA gene sequences PCR amplified from total DNA extracted from the guts and feces collected from larvae of Manduca sexta. Larvae were fed leaf material of either a transgenic (T) or a nontransgenic (N) tobacco cultivar. NL indicates leaves from nontransgenic plants, and TL indicates leaves from transgenic plants. E indicates the total DNA extracted from the eggs of M. sexta and M a SSCP migration marker. The numbers following the indication of the plant cultivar indicate the instar stage (first to fifth) from which samples were collected. For the gut material from the first and second instar stages, DNA was extracted from whole larvae, while for the other instar stages, only the food bolus was used. White triangles within the gel figure indicate the bands which were analyzed by DNA sequencing (see Table 1).
The SSCP profiles from larva in different developmental stages, as well as from gut material or freshly shed feces, were highly similar to each other (Fig. 1). The dominant band 2 was identical to the 16S rRNA gene sequence of three different species from the genus Enterococcus. Band 1, the Burkholderia-Cupriavidus relative, occurred in profiles from the guts and feces, with a tendency to be more abundant in the gut than in feces and also to be more abundant in the guts and feces originating from larvae fed with the nontransgenic leaves. Another inconsistent band (band 4) was identical to the 16S rRNA genes of yet-uncultured bacteria of the order Sphingobacteriales (Bacteroidetes) which had been detected from a variety of environmental samples, including insect guts, rhizospheres, or soils, but with no cultured isolate in its immediate phylogenetic vicinity (Table 1).
SSCP profiles of RT-PCR-amplified partial 16S rRNA sequences.
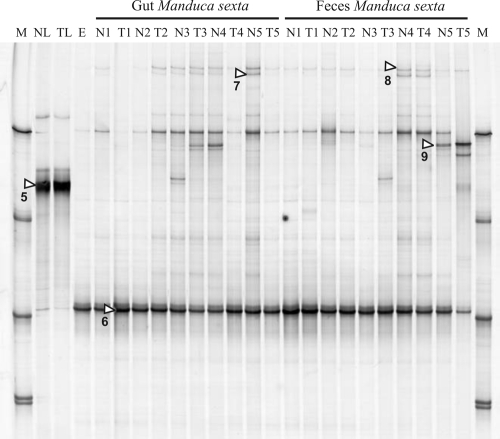
Only one dominant band was visible in the SSCP profiles from the leaves, and sequencing assigned this band to the same Burkholderiales member described above (Table 1). This was expected due to its identical position in both SSCP gels (Fig. 1 and 2). Identity between both sequences was 98.4%, and these differences were mainly caused by some undefined nucleotides of the RNA-derived sequence at the 3′ end (data not shown), suggesting that the real similarity between both PCR products could be even higher. The SSCP profiles from eggs, larvae, and feces were all dominated by only one band (band 6, Fig. 2), and the sequence of this band was identical to the partial 16S rRNA sequence of the Enterococcus members and band 2 described above. One additional, less-dominant band in the position of SSCP marker 1 could not be further characterized, even after several cloning attempts (data not shown). No differences were seen in the profiles of larvae fed with transgenic or nontransgenic leaves. Other less-abundant bands were detected in some profiles from gut material or feces. Three of them were characterized by nucleic acid sequencing, and database comparisons revealed the presence of Caulobacter, Pseudomonas, and Citrobacter sedlakii relatives (Table 1). The C. sedlakii related sequence (band 9) was identical to the sequence detected by the DNA-based analyses of the feces, guts, and eggs (band 3; see above). In the rRNA-based profiles, the C. sedlakii band appeared only in some gut and feces profiles but not from eggs, suggesting that it was metabolically inactive with eggs.
FIG. 2.
SSCP profiles of RT-PCR-amplified 16S rRNA sequences from total RNA extracted from the guts and feces of Manduca sexta larvae. For more details, see the legend to Fig. 1.
Detection of metabolically active bacteria by SIP and SSCP of the RT-PCR-amplified rRNA sequences.
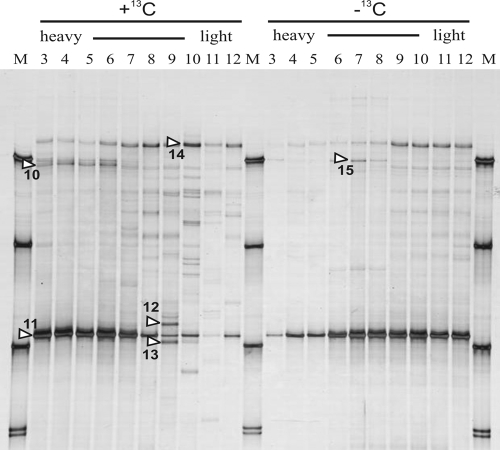
SIP was conducted of the total nucleic acids extracted from the larvae of the second instar stage. Control larvae grew up on tobacco leaves which had been grown in a normal atmosphere, while those selected for SIP grew up on highly 13C-enriched plant material starting at egg hatching. As expected, the SSCP profiles from density gradient fractions of RT-PCR-amplified partial 16S rRNA sequences generally resembled those seen without gradient centrifugation, as previously shown in Fig. 2 (Fig. 3). The single dominant band of all the profiles (band 11) was confirmed to be identical to the sequences of Enterococcus sp. previously detected with the RNA and DNA analyses. In the 12C controls, this band decreased in its intensity with increasing gradient density while it increased in the 13C treatment, demonstrating that it was enriched with 13C and thus derived from metabolically active bacteria. Interestingly, the light-density fractions showed more bands in the 13C treatment than in the 12C control. This was most likely caused by fewer target sequences of Enterococcus being translocated to the high-density fractions, leaving more options for primers to hybridize with DNA sequences from less-abundant bacteria. Bands from less-abundant members (bands 12 to 14) were most closely related to an uncultured Sphingomonas sp., a Flavobacterium sp., and a Delftia acidovorans, respectively (Table 1).
FIG. 3.
SSCP profiles of RT-PCR-amplified 16S rRNA sequences from total RNA extracted from the larvae of the second instar stage using SIP-RNA. Larvae had been fed with the leaves of tobacco plants grown in a 13CO2-atmosphere (+13C) or a control with normal atmospheric CO2 (−13C). Total RNA was separated on trifluoroacetate gradients separating 13C RNA from 12C RNA.
One SSCP band (band 10) was enriched in the high-density fractions of the 13C treatment. This band, identical in its sequence to partial rRNA genes of different Enterobacter spp. (Table 1), was obviously caused by metabolically active bacteria, as it was lacking in the corresponding heavy fractions of the 12C controls. Another sequence isolated from 12C-generated profiles, thus not enriched by 13C metabolism, was identical to the 16S rRNA of a Bacillus licheniformis.
DISCUSSION
The major part of the gut of Manduca sexta larvae, the midgut, provides alkaline conditions (pH > 9) and a negative redox potential, the latter indicating anaerobic conditions (3). In previous studies, we found that ingested food passed through the gut at a high flow rate: for fifth-instar-stage larvae, an average complete gut passage lasted only about 2 h while transporting approximately 1 g of ingested leaf material (wet weight) (5). Cultivation methods suggested that the gut-transported substrate was colonized with bacteria, as approximately 108 bacterial cells were found per g freshly shed feces (5). Detection by cultivation, however, cannot distinguish between metabolically active and inactive cells. The diversity of the potential bacterial carbon and energy sources which come with the ingested leaf material, i.e., cellulose, hemicellulose, pectins, proteins, nicotine, and other alkaloids, could support a diverse bacterial community in the gut. However, despite this diversity of nutrients, the DNA-based SSCP profiles of this study revealed low bacterial diversity, as only one strong and two weak bands were detected. The developmental stages (the first to fifth instar stages) had no clear effect on this bacterial community structure which would have become visible by altered SSCP profiles. However, the quality of the food may have had a minor influence, as a slight difference related to the presence of a Burkholderia relative was detected between the transplastomic and the nonengineered tobacco cultivar. Bacterial communities with highly similar structure were also detected with the SSCP profiles generated from gut contents and feces, indicating that all major gut bacteria were associated with the food bolus and thus also shed. A comparable low diversity of the gut bacteria was also found in analyses of the guts of gypsy moth (Lymantria dispar) larvae which provide similar alkaline conditions (6). In contrast, the guts of larvae with less-alkaline conditions, as they occur, e.g., in beetles (Coleoptera), flies (Diptera), or bees (Apoidea), harbor much higher bacterial diversity (18, 28, 37).
The DNA-based SSCP profiles of this study revealed clear differences between the bacterial communities on leaves, eggs, and guts or feces. The dominant band for leaves was also seen for eggs. In samples from the gut contents and feces, this band occurred at lower relative intensities. Phylogenetic analyses assigned this bacterium to the Burkholderiales (Betaproteobacteria), a group which comprises several bacterial species with a high potential to colonize plant material (4). The relatively lower intensity of this band in gut contents and feces suggests that a population decline took place during the gut passage. For other insects with a similar rod-shaped gut architecture and fast gut passages of ingested feed, like Collembola, it has been demonstrated that gut passages can support bacterial growth or digestion depending on the bacterial species (51). In fact, in this study, RNA-based SSCP profiles indicated that the Burkholderia phylotype was metabolically active on the leaf material but not on eggs, gut material, and feces, where it was seen only in the DNA-based profiles.
DNA-based SSCP profiles from eggs also retrieved two phylotypes, one with 100% sequence identity to C. sedlakii and another with 100% identity to Enterococcus. The C. sedlakii was also, however inconsistently, detected in samples from guts and feces. This band was also seen in the rRNA-based profiles from the gut and fecal material, suggesting that it was metabolically active in the gut but not on eggs where no band was seen in rRNA-based profiles. Members of the genus Citrobacter have already been isolated from different insect guts, suggesting that they are in fact metabolically active gut inhabitants (16, 45). It should be noted that the species C. sedlakii can also be a human pathogen associated with meningitis in premature infants (17). Sequence identity to 16S rRNA genes, however, does not allow the conclusion that the detected bacteria were pathogenic, as avirulent strains have also been described (41).
The Enterococcus phylotype was clearly the most abundant bacterial gut inhabitant, as indicated by the dominant bands in both DNA- and RNA-based SSCP profiles. It was not seen in any SSCP profiles of tobacco leaves, but it occurred with DNA- and RNA-based profiles of eggs, which strongly suggests that it was already metabolically active there. Members of the genus Enterococcus are among the most common gut bacteria detected in larval guts across a diversity of insect orders (35), including Diptera (1, 9, 53) and Lepidoptera (6, 24), but their ecological range for other habitats is also enormous (20, 25, 31, 39, 42). The occurrence of active cells associated with eggs, inside the gut and later on in feces, also points to a versatile metabolic adaptation. Comparison of the rRNA-based profiles from eggs and gut material revealed that the Enterococcus phylotype originated from eggs and not from leaves. The vertical transmission of gut bacteria from mothers to larvae via egg contamination has been described for beetles (Coleoptera) or fruit flies (Diptera) where mothers leave bacteria on the egg surface and larvae pick up these bacteria by licking or feeding on egg shells (11, 13). Data from this study indicate that the same process is also relevant for larvae of M. sexta and possibly lepidopteran larvae in general. In fact, M. sexta larvae typically start to consume their egg cases immediately after hatching (43). Interestingly, the egg itself seemed to support the metabolic activity of the prospective gut inoculant, which may be favorable for efficient gut colonization immediately after hatching. While rRNA-based SSCP profiles were an indication of metabolic activity based on the assumption that active bacteria contain more ribosomal particles than inactive ones, a valuable approach as also demonstrated in other studies (19, 27, 36, 40), SIP of 16S rRNA sequences was direct proof of metabolic activity. Beginning from egg hatching, the larvae of this study grew up with only 13C-labeled carbon sources originating from the tobacco leaves previously cultivated in a 13CO2 atmosphere. Thus, the complete larval biomass was highly enriched by 13C. This did not allow a distinction between bacteria utilizing plant material and others, metabolically active, using other carbon sources excreted into the gut lumen from the insects themselves, i.e., the peritrophic membrane, which consists of chitin and proteins and contains old epithelial cells shed during molting (8, 50). The SIP results of this study confirmed that the Enterococcus phylotype was the metabolically dominant bacterium inside the gut. In addition, an Enterobacter relative was also detected from fractions highly enriched in 13C, demonstrating that SIP was more sensitive to detect the metabolically active bacteria than the “conventional” rRNA profiles. An Enterobacter was also found as a consistent gut colonizer of the gypsy moth (6). Furthermore, additional sequences were retrieved from the 12C-enriched fractions, which were suspected to originate from metabolically less-active or inactive bacteria. The 16S rRNA-based phylogenetic analyses revealed that these sequences were from bacterial species which had already been detected in other lepidopteran larval guts, e.g., Bacillus licheniformis, Flavobacterium sp., and Delftia acidovorans (37, 38, 58). These bacteria were obviously not among the dominant bacteria associated with the food (tobacco leaves) or eggs, as judged by their absence in DNA-based SSCP profiles. We suspect that they were enriched during the gut passage. SIP RNA demonstrated that this enrichment was not caused by active growth, suggesting that they might have been successful in withstanding digestion inside the gut, in contrast to the leaf-associated Burkholderia member described above.
In summary, this study demonstrates that cultivation-independent DNA- and RNA-based SSCP profiling of bacterial SSU rRNA genes, in combination with 13C enrichment and SIP-RNA analyses and phylogenetic analyses, provides a powerful approach for exploring the origin, diversity, and metabolic activity of the bacterial community associated with insect larvae. The methods should have the potential to also explore more complex bacterial communities as detected in this study with M. sexta larvae and to investigate interactions between insects and microorganisms as they take place a billionfold in nature.
Acknowledgments
Karin Trescher supported this work with excellent technical assistance, which we gratefully acknowledge. We thank Jean-Michel Monier, Pascal Simonet, and Timothy M. Vogel, Lyon, Tillmann Lueders, Munich, Germany, and Anja B. Dohrmann, Braunschweig, Germany, for helpful discussions and support.
The work was a part of and financially supported by the EU project TRANSBAC QLK3-2001-02242 (fifth RTD program).
Footnotes
Published ahead of print on 10 October 2008.
REFERENCES
- 1.Ahmad, A., A. Broce, and L. Zurek. 2006. Evaluation of significance of bacteria in larval development of Cochliomyia macellaria (Diptera: Calliphoridae). J. Med. Entomol. 43:1129-1133. [DOI] [PubMed] [Google Scholar]
- 2.Appel, H. M., and L. W. Maines. 1995. The influence of host-plant on gut conditions of gypsy moth (Lymantria-Dispar) caterpillars. J. Insect Physiol. 41:241-246. [Google Scholar]
- 3.Appel, H. M., and M. M. Martin. 1990. Gut redox conditions in herbivorous lepidopteran larvae. J. Chem. Ecol. 16:3277-3290. [DOI] [PubMed] [Google Scholar]
- 4.Berg, G., L. Eberl, and A. Hartmann. 2005. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7:1673-1685. [DOI] [PubMed] [Google Scholar]
- 5.Brinkmann, N., and C. C. Tebbe. 2007. Leaf-feeding larvae of Manduca sexta (Insecta, Lepidoptera) drastically reduce copy numbers of aadA antibiotic resistance genes from transplastomic tobacco but maintain intact aadA genes in their feces. Environ. Biosafety Res. 6:121-133. [DOI] [PubMed] [Google Scholar]
- 6.Broderick, N. A., K. F. Raffa, R. M. Goodman, and J. Handelsman. 2004. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 70:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brune, A. 1998. Termite guts: the world's smallest bioreactors. Trends Biotechnol. 16:16-21. [Google Scholar]
- 8.Chamberlin, M. E., C. M. Gibellato, R. J. Noecker, and E. J. Dankoski. 1997. Changes in midgut active ion transport and metabolism during larval-larval molting in the tobacco hornworm (Manduca sexta). J. Exp. Biol. 200:643-648. [DOI] [PubMed] [Google Scholar]
- 9.Cox, C. R., and M. S. Gilmore. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75:1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deni, J., B. Message, M. Chioccioli, and D. Tepfer. 2005. Unsuccessful search for DNA transfer from transgenic plants to bacteria in the intestine of the tobacco horn worm Manduca sexta. Transgenic Res. 14:207-215. [DOI] [PubMed] [Google Scholar]
- 11.de Vries, E. J., G. Jacobs, and J. A. J. Breeuwer. 2001. Growth and transmission of gut bacteria in the western flower thrips, Frankliniella occidentalis. J. Invertebr. Pathol. 77:129-137. [DOI] [PubMed] [Google Scholar]
- 12.Dohrmann, A. B., and C. C. Tebbe. 2004. Microbial community analysis by PCR-single-strand conformation polymorphism (PCR-SSCP), p. 809-838. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. L. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dodrecht, The Netherlands.
- 13.Douglas, A. E., and C. B. Beard. 1997. Microbial symbiosis in the midgut of insects, p. 315-333. In M. Lehane (ed.), Biology of the insect midgut. Academic Press, New York, NY.
- 14.Douglas, A. E., L. B. Minto, and T. L. Wilkinson. 2001. Quantifying nutrient production by the microbial symbionts in an aphid. J. Exp. Biol. 204:349-358. [DOI] [PubMed] [Google Scholar]
- 15.Dow, J. A. T. 1986. Insect midgut function. Adv. Insect Physiol. 19:187-328. [Google Scholar]
- 16.DugalicVrndic, N. 1997. A study of the microbial flora of the bee intestine. Acta Vet. (Belgrade) 47:135-139. [Google Scholar]
- 17.Dyer, J., K. C. Hayani, W. M. Janda, and P. C. Schreckenberger. 1997. Citrobacter sedlakii meningitis and brain abscess in a premature infant. J. Clin. Microbiol. 35:2686-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egert, M., B. Wagner, T. Lemke, A. Brune, and M. W. Friedrich. 2003. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6659-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felske, A., A. Wolterink, R. VanLis, W. M. de Vos, and A. D. L. Akkermans. 2000. Response of a soil bacterial community to grassland succession as monitored by 16S rRNA levels of the predominant ribotypes. Appl. Environ. Microbiol. 66:3998-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelsomino, R., M. Vancanneyt, T. M. Cogan, S. Condon, and J. Swings. 2002. Source of enterococci in a farmhouse raw-milk cheese. Appl. Environ. Microbiol. 68:3560-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glendinning, J. I. 2002. How do herbivorous insects cope with noxious secondary plant compounds in their diet? Entomol. Exp. Appl. 104(1):15-25. [Google Scholar]
- 22.Goodman, W. G., R. O. Carlson, and K. L. Nelson. 1985. Analysis of larval and pupal development in the tobacco hornworm (Lepidoptera, Sphingidae), Manduca sexta. Ann. Entomol. Soc. Am. 78:70-80. [Google Scholar]
- 23.Herzog, G. A., R. M. McPherson, D. C. Jones, and R. J. Ottens. 2002. Baseline susceptibility of tobacco hornworms (Lepidoptera: Sphingidae) to acephate, methomyl and spinosad in Georgia. J. Entomol. Sci. 37:94-100. [Google Scholar]
- 24.Inglis, G. D., A. M. Lawrence, and F. M. Davis. 2000. Pathogens associated with southwestern corn borers and southern corn stalk borers (Lepidoptera: Crambidae). J. Econ. Entomol. 93:1619-1626. [DOI] [PubMed] [Google Scholar]
- 25.Inglis, G. D., L. J. Yanke, L. M. Kawchuk, and T. A. McAllister. 1999. The influence of bacterial inoculants on the microbial ecology of aerobic spoilage of barley silage. Can. J. Microbiol. 45:77-87. [PubMed] [Google Scholar]
- 26.Jiang, H. B. 2008. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Sci. 15:53-66. [Google Scholar]
- 27.Ka, J. O., Z. Yu, and W. W. Mohn. 2001. Monitoring the size and metabolic activity of the bacterial community during biostimulation of fuel-contaminated soil using competitive PCR and RT-PCR. Microb. Ecol. 42:267-273. [DOI] [PubMed] [Google Scholar]
- 28.Kadavy, D. R., B. Plantz, C. A. Shaw, J. Myatt, T. A. Kokjohn, and K. W. Nickerson. 1999. Microbiology of the oil fly, Helaeomyia petrolei. Appl. Environ. Microbiol. 65:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanost, M. R., H. B. Jiang, and X. Q. Yu. 2004. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 198:97-105. [DOI] [PubMed] [Google Scholar]
- 30.Kay, E., T. M. Vogel, F. Bertolla, R. Nalin, and P. Simonet. 2002. In situ transfer of antibiotic resistance genes from transgenic (transplastomic) tobacco plants to bacteria. Appl. Environ. Microbiol. 68:3345-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kodjo, A., E. Borges, F. Maurin, and Y. Richard. 1996. Contribution to the study of the bacterial flora in genital tracts of healthy snails (Helix aspersa). Rev. Med. Vet. 147:825-830. [Google Scholar]
- 32.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 34.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, J. D., and J. O. Mundt. 1972. Enterococci in insects. Appl. Microbiol. 24:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez, R. J., H. J. Mills, S. Story, and P. A. Sobecky. 2006. Prokaryotic diversity and metabolically active microbial populations in sediments from an active mud volcano in the Gulf of Mexico. Environ. Microbiol. 8:1783-1796. [DOI] [PubMed] [Google Scholar]
- 37.Mohr, K. I., and C. C. Tebbe. 2006. Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ. Microbiol. 8:258-272. [DOI] [PubMed] [Google Scholar]
- 38.Mohr, K. I., and C. C. Tebbe. 2007. Field study results on the probability and risk of a horizontal gene transfer from transgenic herbicide-resistant oilseed rape pollen to gut bacteria of bees. Appl. Microbiol. Biotechnol. 75:573-582. [DOI] [PubMed] [Google Scholar]
- 39.Müller, T., A. Ulrich, E. M. Ott, and M. Müller. 2001. Identification of plant-associated enterococci. J. Appl. Microbiol. 91:268-278. [DOI] [PubMed] [Google Scholar]
- 40.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park, C. H., E. A. Martin, and E. L. White. 1998. Isolation of a nonpathogenic strain of Citrobacter sedlakii which expresses Escherichia coli O157 antigen. J. Clin. Microbiol. 36:1408-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reid, K. C., F. R. Cockerill, and R. Patel. 2001. Clinical and epidemiological features of Enterococcus casseliflavus/flavescens and Enterococcus gallinarum bacteremia: a report of 20 cases. Clin. Infect. Dis. 32:1540-1546. [DOI] [PubMed] [Google Scholar]
- 43.Reinecke, J. P., J. S. Buckner, and S. R. Grugel. 1980. Life-cycle of laboratory-reared tobacco hornworms, Manduca sexta—study of development and behavior, using time-lapse cinematography. Biol. Bull. 158:129-140. [Google Scholar]
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Sanchez, C., F. Hernandez, P. Rivera, and O. Calderon. 1994. Indigenous flora in cockroaches (Dictyoptera, Blattidae and Blattellidae)—a bacteriological and ultrastructural analysis. Rev. Biol. Trop. 42:93-96. [Google Scholar]
- 46.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmalenberger, A., and C. C. Tebbe. 2003. Bacterial diversity in maize rhizospheres: conclusions on the use of genetic profiles based on PCR-amplified partial small subunit rRNA genes in ecological studies. Mol. Ecol. 12:251-261. [DOI] [PubMed] [Google Scholar]
- 48.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steppuhn, A., K. Gase, B. Krock, R. Halitschke, and I. T. Baldwin. 2004. Nicotine's defensive function in nature. PLoS Biol. 2:1074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terra, W. R. 2001. The origin and functions of the insect peritrophic membrane and peritrophic gel. Arch. Insect Biochem. Physiol. 47:47-61. [DOI] [PubMed] [Google Scholar]
- 51.Thimm, T., A. Hoffmann, H. Borkott, J. C. Munch, and C. C. Tebbe. 1998. The gut of the soil microarthropod Folsomia candida (Collembola) is a frequently changeable but selective habitat and a vector for microorganisms. Appl. Environ. Microbiol. 64:2660-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tittiger, C. 2004. Functional genomics and insect chemical ecology. J. Chem. Ecol. 30:2335-2358. [DOI] [PubMed] [Google Scholar]
- 53.Toth, E. M., E. Hell, G. Kovacs, A. K. Borsodi, and K. Marialigeti. 2006. Bacteria isolated from the different developmental stages and larval organs of the obligate parasitic fly, Wohlfahrtia magnifica (Diptera: Sarcophagidae). Microb. Ecol. 51:13-21. [DOI] [PubMed] [Google Scholar]
- 54.Toth-Prestia, C., and I. N. Hirshfield. 1988. Isolation of plasmid-harboring Serratia plymuthica from facultative gut microflora of the tobacco hornworm, Manduca sexta. Appl. Environ. Microbiol. 54:1855-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wernegreen, J. J. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3:850-861. [DOI] [PubMed] [Google Scholar]
- 57.Wink, M., and V. Theile. 2002. Alkaloid tolerance in Manduca sexta and phylogenetically related sphingids (Lepidoptera: Sphingidae). Chemoecology 12:29-46. [Google Scholar]
- 58.Xiang, H., G. F. Wei, S. Jia, J. Huang, X. X. Miao, Z. Zhou, L. P. Zhao, and Y. P. Huang. 2006. Microbial communities in the larval midgut of laboratory and field populations of cotton bollworm (Helicoverpa armigera). Can. J. Microbiol. 52:1085-1092. [DOI] [PubMed] [Google Scholar]