Abstract
Enterobacter cloacae, one of the indigenous gut bacteria of the Formosan subterranean termite (Coptotermes formosanus), was genetically modified with a transposon Tn5 vector containing genes (tcdA1 and tcdB1) encoding orally insecticidal proteins from the entomopathogenic bacterium Photorhabdus luminescens subsp. laumondii TT01, a symbiont of the entomopathogenic nematode Heterorhabditis bacteriophora, for termite control. In the laboratory, termites were fed filter paper inoculated with the recombinant bacteria. The chromosomal expression of the introduced genes showed that there were insecticidal activities against termite workers and soldiers challenged with the transformed bacteria. After termites were fed recombinant bacteria, the termite mortality was 3.3% at day 5, and it increased from 8.7% at day 9 to 93.3% at day 29. All the dead termites contained the recombinant bacteria in their guts. Transfer of the recombinant bacteria occurred between donor workers (initially fed recombinant bacteria) and recipient workers (not fed). More than 20% of the recipient termites ingested recombinant bacteria within 2 h, and 73.3% of them had ingested recombinant bacteria after 12 h. The method described here provides a useful alternative for sustainable control of the Formosan subterranean termite (C. formosanus) and other social insects, such as the imported red fire ant (Solenopsis invicta).
The Formsan subterranean termite (Coptotermes formosanus Shiraki) (Isoptera: Rhinotermitidae) is an ecologically and economically important social insect (26). Although this termite is an excellent food and energy transformer and decomposer in nature, it is a serious destroyer of wood structures built by humans. In China, the cost of this termite species due to damage and control is about $2 billion annually (23). In the United States, the cost of damage by and control of Formosan termites is estimated to be $1 billion annually (22).
This insect pest lives in groups and colonies and engages in social interactions, such as mutual grooming and food sharing. In a social insect colony, the reproductive females are responsible for reproduction. To control an entire termite colony, it is necessary to kill not only the foraging workers and soldiers but also the reproductive females.
Many methods have been tried to eliminate termite populations (32). Chemical control methods are the major choices. However, the toxicants used are associated with certain risks and shortcomings (46). Since certain soil insecticides (chlordane and other cyclodienes) were banned, public demand has increased for reduced-risk, environmentally friendly techniques for termite control (7, 33).
A bait approach using slow-acting toxicants, such as insect growth regulators, is one of the most promising reduced-risk approaches for termite control (32). In this system, the foraging workers consume the bait toxicant, transmit the active ingredient through the colony via food exchange and grooming among nest mates, and die of poisoning. A limitation of current baiting systems is that the concentration of the bait toxicant is diluted after a large number of foragers directly contact and spread the toxicant during social interactions. At the end of the process, the diluted toxicants are not effective against the recipients. If a high concentration of the inoculum is used, the foragers may be killed before they reach the nest.
Theoretically, baiting systems could be improved by employing self-sustaining, self-replicating, and self-perpetuating biological control agents, such as entomopathogenic nematodes, viruses, fungi, and bacteria (18, 27). Although some of these entomopathogens have been used for control of termites in laboratories (7, 14, 37, 41, 44), the results in the field are not as good as expected. The limited success of the biological agents in the field may be due to several factors. For example, the efficacy of most pathogens in their natural state is too low, and the pathogens are not generally persistent in the host environment. In addition, the target insects are able to recognize and avoid contact with pathogens (14), remove them from their nestmates through grooming behavior, and isolate infected individuals from the colony (21). Termites also have an efficient immune system that is effective against foreign pathogenic microbe infections (42, 43). Genetically engineered termite gut bacteria could be candidates for effective termite control (18).
Termites harbor a diverse community of microbes in their guts, including protozoa (Eucarya), archaea, and bacteria (1), and they depend on these microbes for survival. The gut microbes are naturally exchanged between colony members through grooming, food exchange, and coprophagy (20), they provide the nitrogen, carbon, and energy requirements of the termite hosts, and they may protect their termite hosts from invasion by foreign bacteria (8, 36). Using gut bacteria of termites, which can reside within their hosts, as “shuttles” to deliver, express, and spread foreign genes has been attempted with termite colonies (18) and other insects (2, 28, 38, 45). The ice nucleation gene iceA of Erwinia ananas 110 was integrated into the chromosomes of Enterobacter cloacae for possible control of insect pests (35). E. cloacae, which is also one of the dominant bacteria in the termite gut (1), was genetically engineered to express green fluorescent protein (GFP) in termite colonies. The transgenic bacteria could deliver, express, and spread the GFP gene in termite colonies and could establish a persistent population in the termite gut for up to 11 weeks (18). However, no insecticide genes have been introduced into the chromosome of E. cloacae to create a bioinsecticide agent for effective control of termites in a colony.
The goals of the present study were (i) to construct a recombinant transposon (Tn5) vector with genes (tcdA1 and tcdB1) encoding orally insecticidal proteins from the entomopathogenic bacterium Photorhabdus luminescens subsp. laumondii TT01 (Enterobacteriaceae), a symbiont of the entomopathogenic nematode Heterorhabditis bacteriophora (9); (ii) to integrate this transposon into the chromosomal DNA of E. cloacae, one of the major termite gut bacterial species; (iii) to introduce the bacterial shuttle in order to deliver the toxic genes to termite colonies; (iv) to determine the expression of genes encoding oral toxicity against termites in the absence of selective pressure; and (v) to evaluate the bacterial shuttle using some of the criteria and methods defined by Durvasula et al. (10), including ingestion of the bacteria and transfer to the termite gut (method of delivery) and transmission of the transgenic bacteria among workers (method of dispersal).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Strains and plasmids used in this study are listed in Table 1. P. luminescens TT01 was donated by Lihong Qiu of Zhongshan University, Guangzhou, China. Escherichia coli strain S17-1(λpir) was obtained from the Institute of Plant Protection, Chinese Academy of Agriculture Sciences, Beijing, China, and E. cloacae was obtained from Guangdong Microbiological Institute, Guangdong Academy of Sciences, Guangzhou, China. Bacteria were routinely cultured on Luria-Bertani (LB) agar (1.5% [wt/vol] Bacto agar) or in LB broth at 200 rpm and 28°C for P. luminescens and E. cloacae and 37°C for E. coli, unless otherwise indicated. When required, 50 μg/ml of kanamycin (Sigma, United States), 50 μg/ml of disodium carbenicillin (Sigma), 50 μg/ml of ampicillin (Sigma), 50 μg/ml of chloromycetin (Sigma), or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (MD Bio, United States) was added to the medium. All medium components used in this study were purchased from Oxoid Company (Basingstoke, Hampshire, United Kingdom).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or referencea |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | Cloning strain | TaKaRa |
| BL21(DE3) | General purpose expression host | Novagen |
| S17-1(λpir) | E. coli lysogenized with λpir | IPP, CAAS |
| DH5α/pUC19-tcdA1B1 | DH5α with pUC19-tcdA1B1, Ampr | This study |
| BL21(DE3)/pET-32a(+) | BL21(DE3) with pET-32a(+), Carr | This study |
| BL21(DE3)/pET-32a(+)-tcdA1B1-1 | BL21(DE3) with pET-32a(+)-tcdA1B1-1, Carr | This study |
| BL21(DE3)/pET-32a(+)-tcdA1B1-2 | BL21(DE3) with pET-32a(+)-tcdA1B1-2, Carr | This study |
| BL21(DE3)/pET-32a(+)-tcdA1B1 | BL21(DE3) with pET-32a(+)-tcdA1B1, Carr | This study |
| S17-1(λpir)/pMini-Tn5 | S17-1(λpir) with pMini-Tn5, Kanr | This study |
| S17-1(λpir)/pMini-Tn5-tcdA1B1 | S17-1(λpir) with pMini-Tn5-tcdA1B1, Kanr | This study |
| Photorhabdus luminescens TT01 | Phase I variant | 9 |
| Enterobacter cloacae | Recipient, Cmr | GMI, GDAS |
| Enterobacter cloacae tcdA1B1 | Chromosomal DNA with tcdA1B1, Kanr Cmr | This study |
| Plasmid vectors | ||
| pUC19 | Cloning vector, Ampr | TaKaRa |
| pET-32a(+) | Expression vector, Ampr or Carr | Novagen |
| pMini-Tn5 | Transfer vector, Kanr | 15 |
| pUC19-tcdA1B1 | pUC19 with KpnI-BamHI insert containing tcdA1B1, Ampr | This study |
| pET-32a(+)-tcdA1B1-1 | pET-32a(+) with KpnI-BamHI insert containing tcdA1B1, Carr | This study |
| pET-32a(+)-tcdA1B1-2 | pET-32a(+) with XbaI-BamHI insert containing tcdA1B1, Carr | This study |
| pET-32a(+)-tcdA1B1 | pET-32a(+) with NotI insert containing tcdA1B1, Carr | This study |
| pMini-Tn5-tcdA1B1 | pMini-Tn5 with NotI insert containing tcdA1B1, Kanr | This study |
IPP, CAAS, Institute of Plant Protection, Chinese Academy of Agriculture Sciences; GMI, GDAS, Guangdong Microbiological Institute, Guangdong Academy of Sciences.
General molecular techniques.
General molecular techniques were performed as described by Sambrook and Russell (30). Restriction enzymes (TaKaRa, China), T4 ligase (TaKaRa), and calf intestinal alkaline phosphatase (TaKaRa) were used according to the manufacturer's instructions. Plasmids were extracted from the bacteria with a QIAprep spin miniprep kit (Qiagen, The Netherlands). The genomic DNA was isolated using an E.Z.N.A bacterial DNA kit (Omega, United States). When required, DNA fragments were extracted and purified from agarose gels using a QIAEX II gel extraction kit (Qiagen). The standard PCR was performed using a 25-μl reaction mixture, a thermocycler system (Eppendorf, Germany), and the following primers: P1 primers P1-F (5′-ACCATACGCATCGGACAAAC-3′) and P1-R (5′-CGTAGCGGTTATTCACTCTTCT-3′) to amplify the insecticidal tcdA1 and tcdB1 genes; P2 primers P2-F (5′-TAGCATGCGCGGCCGCTTCTTTCCTGCGTTATCCC-3′) and P2-R (5′-GGTCTAGAGCTTGGCGTAATCATGGTC-3′) to provide the sequence of promoter lacZ from plasmid pUC19; and P3 primers P3-F (5′-GGTCTAGAATGTAAAGGCAACACGGATG-3′) and P3-R (5′-GGAAGGACGGAAAGTGGAGA-3′) to amplify the upstream sequence of the tcdA1 gene from the genomic DNA of P. luminescens TT01 (the underlined nucleotides are the SphI, NotI, and XbaI restriction sites, respectively).
Cloning and expression of genomic DNA fragments containing insecticidal genes in E. coli.
To obtain purified target proteins for producing antisera, genomic DNA fragments containing insecticidal genes were expressed in E. coli.
P. luminescens strain TT01 was used to isolate the insecticidal genes tcdA1 and tcdB1 as the sequence of the whole genome was released (9). The target genes tcdA1 and tcdB1 (GenBank accession number NC_005126) are linked together (about 12 kb) (referred to below as the tcdA1B1 genes), so a plasmid library was established to isolate them from genomic DNA.
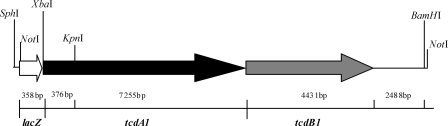
P. luminescens TT01 genomic DNA was digested with KpnI and BamHI. The digested DNA fragments containing a major downstream sequence (7,255 bp) of tcdA1, the entire 4,431-bp sequence of tcdB1, and a partial sequence (2,488 bp) of tccC2 (Fig. 1) were inserted into the vector pUC19 (TaKaRa) and transformed into E. coli strain DH5α. A positive clone was identified by PCR with the P1 primers designed using the tcdA1 and tcdB1 sequences from the NCBI GenBank database. The insert DNA was ligated into pET-32a(+) (Novagen, Germany) and transformed into E. coli strain BL21(DE3). The resulting clone, designated BL21(DE3)/pET-32a(+)-tcdA1B1-1 (Table 1), was identified by PCR performed with the P1 primers. A 638-bp fragment containing a 376-bp upstream sequence of tcdA1 was obtained by amplifying the genomic DNA with the easy-A high-fidelity PCR cloning enzyme (Stratagene, Germany), using the P3 oligonucleotide primers with XbaI and KpnI restriction sites, digested with XbaI and KpnI, and ligated to the corresponding sites of pET-32a(+)-tcdA1B1-1 to generate the whole sequence of tcdA1B1 containing the tcdA1 and tcdB1 genes. The resulting clone, designated BL21(DE3)/pET-32a(+)-tcdA1B1-2 (Table 1), was identified by PCR performed with the P1 primers.
FIG. 1.
Fragment of lacZ-tcdA1B1 which is present in the pET-32a(+)-tcdA1B1 (with SphI and BamHI sites in the vector) and pMini-Tn5 (with NotI sites) constructs expressing tcdA1B1 genes. The tcdA1B1 fragment includes digested DNA containing a 7,255-bp downstream sequence of tcdA1, a 4,431-bp sequence of tcdB1, and a partial sequence (2,488 bp) of tccC2 obtained using KpnI and BamHI, as well as the amplified 376-bp upstream sequence of tcdA1, from P. luminescens TT01 genomic DNA. The enzyme sites indicated are the sites used for subcloning (see text). Sizes are also indicated.
In order to coexpress the tcdA1B1 combined genes in E. coli and E. cloacae, a 358-bp fragment containing the lacZ promoter was amplified from the vector pUC19 using the P2 oligonucleotide primers with SphI and NotI sites in the forward primer and an XbaI site in the reverse primer [XbaI and SphI sites were used for pET-32a(+), and NotI was used for pMini-Tn5] and introduced upstream of the tcdA1B1 genes in pET-32a(+)-tcdA1B1-2 to obtain a fragment containing the lacZ promoter and tcdA1B1. The plasmid was transformed into E. coli stain BL21(DE3). The resulting clone, designated BL21(DE3)/pET-32a(+)-tcdA1B1 (Table 1), was identified by PCR performed with the P1 primers and was induced to express the corresponding proteins using the procedure described in the manufacturer's instructions (pET manual; Novagen). The vector without the insert was transformed into the same E. coli strain as a control.
For expression of the pET-32a(+)-tcdA1B1 product, a single bacterial colony was inoculated into 50 ml LB broth containing carbenicillin (50 μg/ml) and grown at 37°C. When the optical density at 600 nm (OD600) of the culture was 0.6, gene expression was induced by incubation with 1 mM IPTG at 30°C overnight. The occurrence and accumulation of recombinant proteins during culturing were examined by performing 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described below.
Cloning and expression of genomic DNA fragments containing insecticidal genes in E. cloacae.
The fragment containing the lacZ promoter and tcdA1B1 was excised from pET-32a(+)-tcdA1B1 with NotI and cloned into the NotI site of the transposon vector pMini-Tn5 (pUTkm) (15, 45) with a kanamycin resistance marker to obtain pMini-Tn5-tcdA1B1. The plasmid was transformed into E. coli strain S17-1(λpir), a donor bacterium with a transferable gene (tra). The vector without the insert was transformed into the same E. coli strain as a control.
A colony of donor strain E. coli S17-1(λpir) with pMini-Tn5-tcdA1B1 was grown in 50 ml LB broth containing kanamycin (50 μg/ml) at 37°C for 18 h, and then 1/100 of the culture was transferred into the same volume of fresh LB broth. The cultures were incubated at 37°C until the OD600 was 1.0. A 25-ml aliquot was centrifuged (1,000 × g for 10 min at 10°C), and the pellet was washed in 25 ml of a sterile 10 mM MgSO4 solution. The recipient strain was prepared in a similar way. E. cloacae was grown overnight at 28°C in LB broth containing chloromycetin (50 μg/ml) until the OD600 was 1.0. Cells were collected and washed with a 10 mM MgSO4 solution (1,000 × g for 10 min at 10°C). The donor and recipient cell pellets were each resuspended in 1 ml of a 10 mM MgSO4 solution. Then 0.1 ml of the donor culture and 0.5 ml of the recipient culture were mixed and plated on LB plates with 0.2-μm-pore-size nitrocellulose filters. The plates were incubated for 24 h at 28°C. The bacterial lawns growing on the filters were scraped into a 10 mM MgSO4 solution and vortexed to resuspend the cells. Transconjugants were selected by plating these cells on LB agar containing kanamycin (50 μg/ml) and chloromycetin (50 μg/ml) and incubating the preparations at 28°C for 24 h.
The resulting colonies on the selective plates were analyzed by PCR and Southern blotting to confirm that the insecticidal fragment was integrated into the chromosome of E. cloacae. XbaI- and BamHI-digested genomic DNA from the E. cloacae strain with tcdA1B1 genes (designated E. cloacae tcdA1B1) was used for Southern blot analysis. The probes used for the Southern blot analysis were created by amplifying a 778-bp fragment of the tcdA1B1 genes with the P1 primers. Labeling of DNA probes and DNA hybridization were performed by using a DIG High Prime DNA labeling and detection starter kit (Roche, Switzerland) according to the manufacturer's recommendations. In each case, Southern blotting revealed distinct bands corresponding to the correct predicted sizes for the insertion mutant (data not shown).
For expression of the recombinant proteins, a single bacterial colony of E. cloacae tcdA1B1 was inoculated into 50 ml LB broth containing kanamycin (50 μg/ml) and chloromycetin (50 μg/ml) and grown at 28°C for 24 h. The occurrence and accumulation of the TcdA1B1 proteins during culturing were examined by performing 8% SDS-PAGE and Western blotting.
Extraction of TcdA1B1 proteins.
The proteins expressed by E. coli BL21(DE3) containing pET-32a(+)-tcdA1B1 were extracted and purified with the BugBuster reagent, Benzonase nuclease, and rLysozyme solution (Novagen) as described in the pET system manual (10th ed.; Novagen). Briefly, after induction with 1 mM IPTG at 30°C overnight, the bacterial cells were harvested by centrifugation (10,000 × g for 20 min at 4°C) and resuspended in the BugBuster reagent (Novagen) using 5 ml/g (wet weight) pellet. The rLysozyme solution (7.5 kU/g cell paste) and Benzonase nuclease (250 U/g cell paste) were used to treat the suspension on a slow shaker for 30 min at room temperature. Insoluble cell debris obtained by centrifugation (16,000 × g for 20 min at 4°C) was resuspended in the same volume of the BugBuster reagent (see above), incubated with rLysozyme solution (1 kU/ml) at room temperature for 15 min, and centrifuged again. The pellet containing the expressed proteins detected by SDS-PAGE was washed four times with the BugBuster reagent diluted 1/10. The pellet was resuspended in one-half the original culture volume of diluted BugBuster reagent after each wash. The last pellet with most of the expressed proteins was collected by centrifugation (16,000 × g for 20 min at 4°C) and stored at −20°C. The resulting proteins were used to raise antibodies in rabbits.
Extraction of intracellular proteins, including extraction of TcdA1B1 from a P. luminescens TT01 culture, was performed as follows to produce an antigen for an indirect enzyme-linked immunosorbent assay (ELISA). After 48 h of incubation at 28°C, bacterial cells were harvested by centrifugation (10,000 × g for 20 min at 4°C) and then washed three times using phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4; pH 7.4) and centrifugation (10,000 × g for 15 min at 4°C). The pellets were resuspended in PBS and sonicated (at 18 Ω for 20 s) on ice. The supernatants were collected by centrifugation (10,000 × g for 20 min at 4°C) and passed through 0.45-μm filters (Millipore, United States). (NH4)2SO4 was added to the resulting supernatants until the level of saturation was 85%, and the mixed solutions were incubated without shaking at 4°C for 12 h to extract the proteins. After centrifugation (10,000 × g for 20 min at 4°C), each precipitate with extracted proteins was dissolved with sterile distilled water, dialyzed in sterile distilled water at 4°C for 48 h (the sterile distilled water was replaced at 4-h intervals), and then stored at −20°C.
Production of antiserum and indirect ELISA.
Antibodies were raised against purified TcdA1B1 extracts from recombinant E. coli BL21(DE3). New Zealand White rabbits were immunized with TcdA1B1 extracts using subcutaneous injection. Freund's complete adjuvant and incomplete adjuvant (Sigma) were used for the primary and secondary injections, respectively. Prior to emulsification in adjuvant, TcdA1B1 extracts were suspended in PBS. For additional immunizations, purified TcdA1B1 extracts suspended in PBS were used directly without any adjuvant. The quality of the antibodies in sera was monitored by performing ELISA. A total of 2 ml of blood was obtained from the large vein in the center of an ear before every immunization at weekly intervals. As the antigen for ELISA, purified P. luminescens TT01 intracellular proteins were diluted in carbonate buffer (0.1 M Na2CO3, 0.1 M NaHCO3; pH 9.6) at 37°C for 1 h and at 4°C overnight. The ELISA was performed using 1 h of blocking with PBS (pH 7.4) containing 0.05% Tween 20 and 10% bovine serum albumin, 1 h of incubation with diluted antisera, and 1 h of incubation with anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (1/10,000 dilution; Zhongshan Co., Beijing, China). Three 3-min washes with PBS (pH 7.4) containing 0.05% Tween 20 and 10% bovine serum albumin between the steps described above were included. Using 3,3,5,5-tetramebenaidine as the color substrate, the OD450 was determined with a model 550 microplate reader (Bio-Rad, United States). The OD450 of the antiserum increased and was >3.0 after five injections. The antiserum was stored at −80°C in aliquots or was used without further purification.
Gel electrophoresis and Western blot analysis.
To analyze P. luminescens TT01 and recombinant E. cloacae bacterial cells, 8% SDS-PAGE was performed using the standard procedures. Western blotting was performed using polyvinylidene difluoride membranes (Immobilon-PSQ; Millipore, Bedford, MA) and 25 mM Tris-192 mM glycine-20% (vol/vol) methanol. The gels were electroblotted for 3 h at 250 mA (constant current) in a tank blotter (Liuyi, China). Each membrane was processed at room temperature in Tris-buffered saline (50 mM Tris, 150 mM NaCl; pH 7.5) using three 10-min washes between steps. The steps used were overnight blocking with 5% bovine serum albumin in Tris-buffered saline containing 0.1% Tween 20, 2 h of incubation with the antiserum (1/3,000 dilution) at 4°C, and 1 h of incubation with the anti-rabbit immunoglobulin G-alkaline phosphatase conjugate (1/30,000 dilution; Sigma). The blots were visualized using a nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolylphosphate dye kit (Sabc, Shanghai, China).
Stability of the tcdA1B1 genes in E. cloacae.
A colony of recombinant strain E. cloacae tcdA1B1 was inoculated into 5 ml LB broth containing kanamycin (50 μg/ml) and chloromycetin (50 μg/ml) and incubated at 200 rpm and 28°C for 24 h. Then 0.1 ml of the culture was subcultured in the same volume of LB broth without antibiotics every 12 h 10 times. The final culture was spread onto LB agar plates without any antibiotics after it was serially diluted. One hundred resulting colonies were randomly selected and plated on LB agar plates with kanamycin (50 μg/ml) and chloromycetin (50 μg/ml), and the resulting colonies were examined to determine the stability of E. cloacae tcdA1B1 without selective pressure. The final culture was also analyzed by using SDS-PAGE and Western blotting to examine the expression of the inserted genes.
Insecticidal assays.
C. formosanus termite workers and soldiers were collected from cardboard bait buried near trees on the campus of Zhongshan University, Guangzhou, China. Before they were used, freshly collected termites were kept in a large plastic container (diameter, 30 cm; height, 40 cm) in the laboratory at room temperature (25 to 30°C) for 1 week to check for the possible presence of pathogens in the colony. The container was filled with pine (Pinus sp.) wood stakes as food for the termites. To grow liquid bacterial cultures to feed the termites, genetically modified E. cloacae tcdA1B1 was cultured in 50 ml LB broth at 28°C for 16 h without any antibiotics to avoid toxic effects on the termites. The number of E. cloacae tcdA1B1 cells per ml was determined by serially diluting the bacterial cultures with LB broth and plating the dilutions on selective LB agar plates. For oral bioassays, 500-μl portions of bacterial cultures containing about 5.0 × 108 CFU were applied to petri dishes (100 by 15 mm) containing one layer of filter paper (diameter, 9 cm; Sanhuan Product, China). Termites (45 workers and five soldiers per dish) were placed in the petri dishes and incubated at 28°C for 29 days so that they could feed on the filter paper inoculated with the recombinant bacterial culture. Controls were fed filter paper treated with E. cloacae carrying a transposon without the tcdA1B1 genes, with E. cloacae lacking a transposon, or with 500 μl sterile distilled water. Each treatment consisted of three replicates. The dishes were observed every day, and dead individuals were removed and saved at 4°C for at most 3 days to determine the presence of the E. cloacae recombinant in the termite gut when necessary. The percentages of termite mortality were recorded.
Presence of the E. cloacae recombinant in the termite gut.
The dead termites collected in the insecticidal assays were examined to determine the presence of the recombinant E. cloacae tcdA1B1 by carefully sterilizing their surfaces with 70% ethanol for 10 min (18) and removing and crushing their whole guts aseptically in LB agar containing kanamycin (50 μg/ml) and chloromycetin (50 μg/ml). After incubation at 28°C for 24 h, the presence or absence of recombinant bacteria was confirmed first by observing the colony morphology and then by selecting 10 termite gut bacterial colonies and amplifying the fragments of the tcdA1B1 genes by performing PCR with the P1 primers.
Transfer of the E. cloacae recombinant in a termite colony.
We examined whether when the recombinant E. cloacae tcdA1B1 was used in bait systems, the bacteria could be transferred from infected termites (donors) to new termites (recipients). Transfer of the recombinant E. cloacae tcdA1B1 in a termite colony was determined using the modified method of Husseneder and Grace (18).
To distinguish between donor termites and recipients, the donors were dyed by feeding them filter paper containing 1% (wt/wt) Sudan Red 7B (6.0 mg stain per filter; Amresco, United States) for 1 week. It was demonstrated previously that this dye had no influence on the ingestion, spread, or stability of E. cloacae in termites (18). Seven hundred worker donors from one colony were fed 500 μl of recombinant bacteria (106 CFU/μl) for 2 days using the method described above for the insecticidal assays. Ten donors were randomly selected to ensure that they contained recombinant bacteria before the transfer experiment was begun. The recipients were maintained under the same conditions as the donors, but they were fed filter paper treated with 500 μl of sterile distilled water. Two experiments were conducted as follows.
In the first experiment, the transfer of recombinant bacteria between workers was examined every 2 h for 12 h. Fifty donors were combined with 50 recipients in a petri dish (100 by 15 mm), and for the first 12 h, guts were extirpated every 2 h from five recipients and the fragment of the tcdA1B1 genes was amplified by PCR with the P1 primers to determine the presence of recombinant bacteria. Three replicates were used for each treatment.
In the second experiment the transfer of bacteria between workers was examined daily for 6 days with different donor/recipient ratios (1:1, 1:10, and 1:50). For each donor/recipient ratio a total of 100 workers were placed on moist filter paper in petri dishes (100 by 15 mm). The guts of five living recipients and all dead termites were extirpated daily to determine the percentage of termites containing recombinant bacteria. The mortality of the donors and recipients was recorded daily. Three replicates were used for each treatment.
Statistical analysis.
Data expressed as percentages were normalized using arcsine square root transformation and were analyzed using repeated-measures analysis of variance with the Bonferroni adjusted post hoc pair test, using SPSS statistical software. If Mauchly's test of sphericity showed no significant difference in the repeated-measures data (P > 0.05), normal one-way analysis of variance was used and the significance of differences between treatments in each experiment was evaluated by using Duncan's multiple-range test. The values were expressed as means ± standard deviations. A P value of <0.05 was considered statistically significant.
RESULTS
Construction and expression of the tcdA1B1 genes in E. coli and E. cloacae.
The 14-kb KpnI and BamHI fragment from the genomic DNA of P. luminescens TT01 was ligated with a PCR fragment digested by XbaI and KpnI from the same genomic DNA to generate a large fragment containing the tcdA1B1 genes. The lacZ promoter with corresponding SphI, NotI, and XbaI sites was added to control this large fragment properly (Fig. 1). The whole cassette was cloned with suitable restriction sites into the expression vector pET-32a(+) to create pET-32a(+)-tcdA1B1 and into the transposon vector pMini-Tn5 to obtain pMini-Tn5-tcdA1B1.
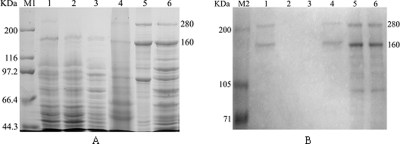
pET-32a(+)-tcdA1B1 was transformed into E. coli strain BL21(DE3), and intracellular proteins in recombinant BL21(DE3) cells induced with IPTG overnight were separated by 8% SDS-PAGE. Two prominent bands at about 280 and 160 kDa were detected for the induced recombinant bacterial cells but not for the control cells (E. coli strain DE3) transformed with pET-32a(+) without an insert (data not shown).
By using conjugation, the pMini-Tn5-tcdA1B1 construct in E. coli strain S17-1(λpir) (chromosomal DNA with the tra gene) was integrated into the chromosomal DNA of E. cloacae, which does not contain the pir gene. Plasmid isolation and PCR identification confirmed the absence of foreign plasmids in the recombinant E. cloacae tcdA1B1. Southern blot analysis of the genomic DNA of the target bacteria with the tcdA1B1 insertion, probed with the fragment of the tcdA1B1 genes, revealed only one hybridization band, which suggested that there was a single insertion pattern for tcdA1B1 in the chromosome of E. cloacae (data not shown). The protein expression in cultures of the recombinant E. cloacae tcdA1B1 was confirmed by 8% SDS-PAGE (Fig. 2). Two prominent bands at about 280 and 160 kDa were obtained for the recombinant bacterial cells, and they were the same sizes as the TcdA1B1 bands obtained for P. luminescens TT01 cells (Fig. 2) but not for the control cells (E. cloacae carrying a transposon without the tcdA1B1 insertion and the wild-type strain of E. cloacae). The sizes of the expressed protein bands are also in accordance with the calculated protein sizes.
FIG. 2.
SDS-PAGE analysis (8% gel) of TcdA1B1 proteins (A) and Western blot analysis of the same samples with TcdA1B1 polyclonal antisera (B). Lane M1, TaKaRa narrow-range protein molecular mass standards (44.3 to 200 kDa); lane M2, Hou-Bio prestained SDS-PAGE standards (2 to 200 kDa); lane 1, E. cloacae cells carrying a transposon with the tcdA1B1 genes; lane 2, E. cloacae cells carrying a transposon without the tcdA1B1 genes; lane 3, E. cloacae cells lacking a transposon; lane 4, P. luminescens TT01 cells; lane 5, recombinant TcdA1B1 proteins extracted from an E. coli BL21(DE3)/pET-32a(+)-tcdA1B1 culture induced with IPTG; lane 6, E. coli BL21(DE3)/pET-32a(+)-tcdA1B1 culture induced with IPTG.
Immunological analysis.
Using the method described in the pET system manual, target proteins were detected in the insoluble cytoplasmic fraction of the cells but not in the medium fraction, the periplasmic fraction, or the soluble cytoplasmic fraction. Recombinant TcdA1B1 proteins were purified by repeated centrifugation and washing using the BugBuster reagent. These proteins were used to produce the polyclonal antisera. The extracted intracellular proteins of P. luminescens TT01 were used as the antigen for ELISA. After four immunizations, the OD450 for sera in ELISA reactions clearly increased. Bleeding was performed after the rabbits were inoculated six times, and the titers of the TcdA1B1 antisera reached stable high levels as determined by ELISA.
Polyclonal antisera raised against extracted proteins that included TcdA1B1 from pET-32a(+)-tcdA1B1 were used to analyze the immunological cross-reactivity of the expressed TcdA1B1 proteins. Western blot analysis of the cell lysates of E. cloacae tcdA1B1 and P. luminescens TT01 probed with TcdA1B1 antisera showed that there were strong reactions with the target proteins (Fig. 2).
Stability of the inserted genes in E. cloacae.
The E. cloacae recombinant was subcultured 10 times without selective antibiotic pressure. SDS-PAGE and Western blotting confirmed that the inserted genes were present and expressed (data not shown).
Insecticidal activity of recombinant E. cloacae against termites.
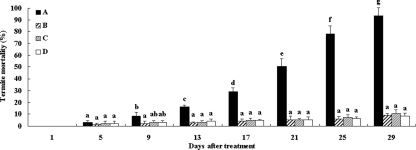
After termites were fed the recombinant bacteria, the mortality was less than 3.3% at day 5, and it increased from 8.7% at day 9 to 93.3% at day 29. However, the average termite mortality for the three controls during the test period was less than 10.7% (Fig. 3). The repeated-measures analysis of variance revealed that there was a significant main effect of time (F = 91.87, P < 0.01), a significant main effect of bacterial treatment (F = 85.44, P < 0.01), and a significant bacterial treatment-time interaction (F = 31.51, P < 0.01). There was a significant difference in the mortalities of the termites treated with recombinant bacteria over time, but there was not a significant effect on the control termite mortalities. To examine the effect of the recombinant bacteria on termite mortality, the mortality data for the treated termites and controls obtained for each time interval were analyzed. The results showed that there were no significant differences in mortality between the treated termites and the controls on day 5; however, the levels of mortality of the termites challenged with the recombinant bacteria were significantly higher than the levels of mortality of the controls starting at day 9, and the level of mortality reached 93.3% at day 29 (Fig. 3). We therefore concluded that the heterologous expression of the cloned tcdA1B1 genes in the recombinant E. cloacae strain resulted in production of recombinant toxins with insecticidal activity against termites.
FIG. 3.
Insecticidal activities of E. cloacae harboring the tcdA1B1 genes against the Formosan subterranean termite. Termites were exposed to the bacterial cultures for 29 days. A, E. cloacae harboring tcdA1B1 genes; B, E. cloacae harboring a transposon without any inserted genes; C, E. cloacae wild-type strain; D, double-distilled water control. The data are averages for three replicates. The error bars indicate standard deviations. Post hoc t tests were performed for each day. For each day bars labeled with different letters are significantly different (P < 0.05).
Presence of the recombinant E. cloacae in the termite gut.
Termite workers ingested the recombinant bacteria rapidly. Colony morphology and PCR were used to confirm that 100% of the collected dead termite workers contained the recombinant bacteria. The results demonstrated that the recombinant bacteria were ingested by termite workers and present in all dead termite workers.
Transfer of recombinant E. cloacae in a termite colony.
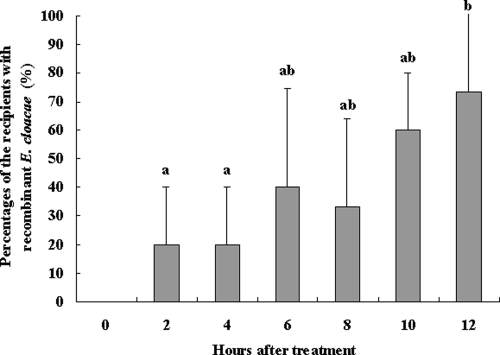
The results of the first transfer experiment showed that in the first 12 h after donors and recipients (50:50) were combined, the rate of transfer of recombinant E. cloacae between workers increased. As soon as 2 h after the donors and recipients were combined, an average of 20% of the recipients contained E. cloacae in their guts. After 12 h, the percentage of recipients containing introduced E. cloacae was 73.3% (Fig. 4). This indicated that the donors transferred the recombinant bacteria to the recipients in a short time, at least under laboratory conditions.
FIG. 4.
Transfer of the recombinant E. cloacae strain between workers in the first 12 h: percentages of the recipients containing recombinant E. cloacae after they were combined with donors. The data are the averages for three replicates. The error bars indicate standard deviations. Bars labeled with different letters are significantly different (P < 0.05).
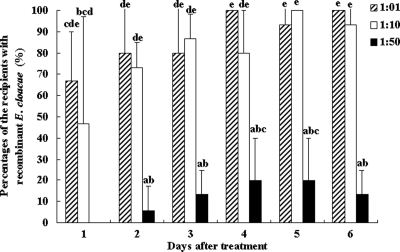
In the second transfer experiment bacterial transfer between workers was measured using three donor/recipient ratios (1:1, 1:10, and 1:50). All of the donors contained recombinant bacteria in their guts after feeding on the recombinant bacteria. When ratios of 1:1 and 1:10 were used, the donors passed the bacteria to 66.7 and 46.7% (averages) of the recipients, respectively, on the first day and to 90 to 100% of the recipients after 6 days. There were no significant differences in the transfer rates for the 1:1 and 1:10 donor/recipient ratios after 6 days. However, when the 1:50 ratio was used, no recombinant bacteria were detected in the recipients on the first day, and on the second day donors passed the bacteria to 5.8% of the recipients. The average proportion of the recipients with recombinant bacteria was 20% after 6 days when the 1:50 ratio was used. Although the transfer rate with the 1:50 ratio was low, a ratio of 1 donor to 50 recipients could still spread recombinant bacteria throughout the laboratory colony (Fig. 5).
FIG. 5.
Transfer of recombinant E. cloacae among workers with different donor/recipient ratios (1:1, 1:10, and 1:50): percentages of recipients containing recombinant E. cloacae. The data are the averages for three replicates. The error bars indicate standard deviations. Mauchly's test of sphericity showed that there were no significant differences in the repeated-measures data (P > 0.05), so a normal one-way analysis of variance was used. Bars labeled with different letters are significantly different (P < 0.05).
In the second experiment, after 6 days the maximum levels of termite mortality were 8% for the donors and 3.3% for the recipients when the ratio was 1:1, 10% for the donors and 2.2% for the recipients when the ratio was 1:10, and 16.7% for the donors and 1.3% for the recipients when the ratio was 1:50. Based on these results, it seemed that most of infected termites were not killed in 6 days, although the donors could spread the bacteria to the recipients quickly. All the dead termites contained recombinant bacteria in their guts.
DISCUSSION
The environmental concerns associated with the use of highly toxic chemical termiticides are forcing the pesticide industry to search for less toxic termite management methods. A biological method is one of the best alternatives (7, 33). To the best of our knowledge, this is the first report showing that insecticidal toxin genes from the symbiotic Photorhabdus bacteria of entomopathogenic nematodes were integrated into the chromosomal DNA of E. cloacae by a transposon method, and the transgenic E. cloacae showed very promising potential for biological control of the Formosan subterranean termite (C. formosanus). The success of the present method provides a useful alternative for sustainable control of termites and other social insects, such as the imported red fire ant (Solenopsis invicta).
E. cloacae was chosen as a shuttle bacterium for termites because it has been isolated from various insect species and has been shown to grown and be maintained in the guts of these insects, including termites (1, 18, 34). Although foreign genes can be efficiently expressed in E. coli, this bacterium survived less than 1 week in the termite gut (17). Bacteria derived from the indigenous gut flora of termites do not trigger defensive or immune responses and adapt well to the physiological and biochemical conditions and selective pressures in the termite gut (18). E. cloacae was transformed previously with an ice nucleation gene to successfully reduce the cold hardiness of the mulberry pyralid moth (Glyphodes pyloalis) (38, 45).
Photorhabdus bacteria belonging to the Enterobacteriaceae are symbionts of entomopathogenic Heterorhabditis nematodes. These nematodes are used as a commercial bioinsecticide for many economically important insect pests (11, 13). The symbiotic bacteria exhibit insecticidal activities against different insects (3, 4, 6, 12, 25). Several insecticidal toxins from Photorhabdus bacteria are involved, especially toxin complex (Tc) proteins (3, 39). The direct use of Photorhabdus bacteria as a biopesticide is severely limited, but the insecticidal Tc proteins expressed in E. coli and in transgenic plants (24) have exhibited oral insecticidal activity against Costelytra zealandica (16), Pieris brassicae, Plutella xylostella, Phaedon cochleariae (25), Manduca sexta (24, 39), and Galleria mellonella (19). Heterologous expression of multiple P. luminescens Tc proteins and of Tc homologues suggests that three components are required to achieve full toxicity: a tcdA-like component, a tcdB-like component, and a tccC-like component (16, 31, 39, 40). However, Pinheiro and Ellar reported that the expression conditions are crucial for observing toxicity (29). In this study, we showed that coexpression of P. luminescens tcdA1 and tcdB1 in E. cloacae is toxic to the subterranean termite C. formosanus when the transgenic bacteria are ingested by the termites and that insecticidal proteins from symbionts of entomopathogenic nematodes are ideal toxic gene products for use against termites.
Rapid ingestion and stable persistence of transgenic bacteria through feeding, as demonstrated not only with subterranean termites (18) but also with other solitary insect species (5, 8, 28), promote the application of transgenic bacteria for control of termite colonies. Although about 73% of the living termite workers had the challenge recombinant bacteria in their guts after 12 h, they were not killed quickly. As shown in Fig. 3, less than 10% of the workers were dead after 9 days, which provided the opportunity to spread the introduced bacteria in the termite colony by social interactions. The transgenic E. cloacae cells were transferred from donors to recipients quickly and efficiently. By using a bait system, it is possible to spread foreign genes throughout a termite field colony for termite control. In the laboratory, even with 1 donor per 50 recipients, spread of the recombinant bacteria to at least 20% of the recipients occurred in 6 days. As indicated by Husseneder and Grace (18), the key to the rapid spread of bacteria to a large number of termite individuals probably depends on the multiplicative effect of interactions of recipients that become secondary donors. However, a reasonable ratio of treated donors to recipients for efficient control of a termite colony in the field is still not known.
For future release of genetically modified bacteria for field application, it is imperative to gather detailed data on the potential environmental impact of the organisms, including the persistence of the transgenic bacteria in the soil and possible gene transfer between bacterial strains. We are currently conducting research to collect detailed data. Based on the present information, it seems that the recombinant E. cloacae bacteria with Tc genes are safe in the environment. First, it has been reported that a transformed E. cloacae strain did not accumulate over time in soil and that the GFP plasmid in the transformed bacteria was not transferred to other soil bacteria (18). In this study, although the pMini-Tn5 expression vector exhibits broad-host-range conjugal mobilization, it is not transposable when it integrates into the chromosomal DNA of a host bacterium due to the lack of the tnp gene in the chromosome (15, 45). Thus, it is believed that the Tn5 transposon in the transgenic E. cloacae strain would also not be transmitted to soil bacteria. Second, the insecticidal genes used here, which were isolated from symbionts of entomopathogenic nematodes and were used in a transgenic plant, are considered promising alternatives to Bacillus thuringiensis genes for management of resistance in a variety of crops (24). Third, the risk of introducing transgenic bacteria into the environment where termites live can be greatly reduced by formulation of proper baits, such as baits which attract the termites but not other insects. Nevertheless, more detailed information is required to evaluate the environmental risk of the genetically modified bacteria for field application.
Acknowledgments
This work was supported by the National Hatch Project for Agriculture, MOST, China (grant 2006GB2E000222), and by the Provincial Natural Foundation of Guangdong (grant 2007-6-7003403).
Footnotes
Published ahead of print on 3 October 2008.
REFERENCES
- 1.Adams, L., and R. Boopathy. 2005. Isolation and characterization of enteric bacteria from the hindgut of Formosan termite. Bioresour. Technol. 96:1592-1598. [DOI] [PubMed] [Google Scholar]
- 2.Bextine, B., C. Lauzon, S. Potter, D. Lampe, and T. A. Miller. 2004. Delivery of a genetically marked Alcaligenes sp. to the glassy-winged sharpshooter for use in a paratransgenic control strategy. Curr. Microbiol. 48:327-331. [DOI] [PubMed] [Google Scholar]
- 3.Bowen, D. J., and J. C. Ensign. 1998. Purification and characterization of a high-molecular-weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl. Environ. Microbiol. 64:3029-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowen, D. J., and J. C. Ensign. 2001. Isolation and characterization of intracellular protein inclusions produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl. Environ. Microbiol. 67:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapco, W., and R. A. Kelln. 1994. Persistence of ingested bacteria in the grasshopper gut. J. Invertebr. Pathol. 64:149-150. [Google Scholar]
- 6.Ciche, T. A., C. Darby, R. U. Ehlers, S. Forst, and H. Goodrich-Blair. 2006. Dangerous liaisons: the symbiosis of entomopathogenic nematodes and bacteria. Biol. Control 38:22-46. [Google Scholar]
- 7.Culliney, T. W., and J. K. Grace. 2000. Prospects for the biological control of subterranean termites (Isoptera: Rhinotermitidae), with special reference to Coptotermes formosanus. Bull. Entomol. Res. 90:9-21. [PubMed] [Google Scholar]
- 8.Dillon, R. J., and V. M. Dillon. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49:71-92. [DOI] [PubMed] [Google Scholar]
- 9.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, and J. F. Charles. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 10.Durvasula, R. V., R. K. Sundaram, C. Cordon-Rosales, P. Pennington, and C. B. Beard. 2003. Rhodnius prolixus and its symbiont, Rhodococcus rhodnii: a model for paratransgenic control of disease transmission, p. 83-95. In K. Bourtzis and T. A. Miller (ed.), Insect symbiosis. CRC Press, Boca Raton, FL.
- 11.Ehlers, R. U., and H. M. T. Hokkanen. 1996. Insect biocontrol with non-endemic entomopathogenic nematodes (Steinernema and Heterorhabditis spp.): conclusions and recommendations of a combined OECD and COST workshop on scientific and regulatory policy issues. Biocontrol Sci. Technol. 6:291-293. [Google Scholar]
- 12.ffrench-Constant, R., N. Waterfield, P. Daborn, S. Joyce, H. Bennett, C. Au, A. Dowling, S. Boundy, S. Reynolds, and D. Clarke. 2003. Photorhabdus: towards a functional genomic analysis of a symbiont and pathogen. FEMS Microbiol. 26:433-456. [DOI] [PubMed] [Google Scholar]
- 13.Georgis, R. 2002. The Biosys experiment: an insider's perspective, p. 357-372. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Wallingford, United Kingdom.
- 14.Grace, J. K. 1995. Microbial termite control, p. 166-167. In Hawaii agriculture: positioning for growth. CTAHR Proceedings. University of Hawaii, Honolulu.
- 15.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurst, M. R., T. R. Glare, T. A. Jackson, and C. W. Ronson. 2000. Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J. Bacteriol. 182:5127-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husseneder, C., J. K. Grace, and D. E. Oishi. 2005. Use of genetically engineered bacteria (Escherichia coli) to monitor ingestion, loss and transfer of bacteria in termites. Curr. Microbiol. 50:119-123. [DOI] [PubMed] [Google Scholar]
- 18.Husseneder, C., and J. K. Grace. 2005. Genetically engineered termite gut bacteria (Enterobacter cloacae) deliver and spread foreign genes in termite colonies. Appl. Microbiol. Biotechnol. 68:360-367. [DOI] [PubMed] [Google Scholar]
- 19.Joo Lee, P., J. Y. Ahn, Y. H. Kim, S. W. Kim, J. Y. Kim, J. S. Park, and J. Lee. 2004. Cloning and heterologous expression of a novel insecticidal gene (tccC1) from Xenorhabdus nematophilus strain. Biochem. Biophys. Res. Commun. 319:1110-1116. [DOI] [PubMed] [Google Scholar]
- 20.La Fage, J. P., and W. L. Nutting. 1978. Nutrient dynamics of termites, p. 165-232. In M. V. Brian (ed.), Production ecology of ants and termites. Cambridge University Press, Cambridge, United Kingdom.
- 21.Lai, P. Y. 1977. Biology and ecology of the Formosan subterranean termite, Coptotermes formasanus, and its suspectibility to the entomogenous fungi, Beauveria bassiana and Metarrhizium anisopliae. Ph.D. thesis. University of Hawaii, Honolulu.
- 22.Lax, A. R., and W. L. A. Osbrink. 2003. United States Department of Agriculture-Agriculture Research Service research on targeted management of the Formosan subterranean termite Coptotermes formosanus Shiraki (Isoptera: Rhinotermitidae). Pest Manag. Sci. 59:788-800. [DOI] [PubMed] [Google Scholar]
- 23.Lin, S. Q. 1987. Present status of Coptotermes formosanus and its control in China, p. 31-36. In M. Tamashir and N. Y. Su. (ed.), Biology and control of the Formosan subterranean termite. College of Tropical Agriculture, University of Hawaii, Honolulu.
- 24.Liu, D., S. Burton, T. Glancy, Z. S. Li, R. Hampton, T. Meade, and D. J. Merlo. 2003. Insect resistance conferred by 283-kDa Photorhabdus luminescens protein TcdA in Arabidopsis thaliana. Nat. Biotechnol. 21:1222-1228. [DOI] [PubMed] [Google Scholar]
- 25.Morgan, J. A. W., M. Sergeant, D. Ellis, M. Ousley, and P. Jarrett. 2001. Sequence analysis of insecticidal genes from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 67:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulrooney, J. E., and P. D. Gerard. 2007. Toxicity of fipronil in Mississippi soil types against Reticulitermes flavipes (Isoptera: Rhinotermitidae). Sociobiology 50:63-70. [Google Scholar]
- 27.Myles, T. G. 2004. Termite control in Canada. Centre Urban Community Stud. Univ. Toronto Res. Bull. 23:4. [Google Scholar]
- 28.Peloquin, J. J., C. R. Lauzon, S. Potter, and T. A. Miller. 2002. Transformed bacterial symbionts re-introduced to and detected in host gut. Curr. Microbiol. 45:41-45. [DOI] [PubMed] [Google Scholar]
- 29.Pinheiro, V. B., and D. J. Ellar. 2007. Expression and insecticidal activity of Yersinia pseudotuberculosis and Photorhabdus luminescens toxin complex proteins. Cell. Microbiol. 9:2372-2380. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2002. Molecular cloning: a laboratory manual, 3rd ed. China Science Press, Beijing, China. (In Chinese.)
- 31.Sergeant, M., P. Jarrett, M. Ousley, and J. A. W. Morgan. 2003. Interactions of insecticidal toxin gene products from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 69:3344-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su, N. Y., and R. H. Scheffrahn. 1998. A review of subterranean termite control practices and prospects for integrated pest management program. Integr. Pest Manag. Rev. 3:1-13. [Google Scholar]
- 33.Sun, J. Z., J. R. Fuxa, and G. Henderson. 2002. Sporulation of Metarhizium anisopliae and Beauveria bassiana on Coptotermes formosanus and in vitro. J. Invertebr. Pathol. 81:78-85. [DOI] [PubMed] [Google Scholar]
- 34.Tanada, Y., and H. K. Kaya. 1993. Insect pathology, p. 28. Academic Press, New York, NY.
- 35.Tang, C. R., F. Z. Sun, X. J. Zhang, T. C. Zhao, and J. Y. Qi. 2004. Transgenic ice nucleation-active Enterobacter cloacae reduces cold hardiness of corn borer and cotton bollworm larvae. FEMS Microbiol. Ecol. 51:79-86. [DOI] [PubMed] [Google Scholar]
- 36.Veivers, P. C., R. W. O'Brien, and M. Slaytor. 1982. Role of bacteria in maintaining the redox potential in the hindgut of termites and preventing the entry of foreign bacteria. J. Insect Physiol. 28:947-951. [Google Scholar]
- 37.Wang, C. L., and J. E. Powell. 2004. Cellulose bait improves the effectiveness of Metarhizium anisopliae as a microbial control of termites (Isoptera: Rhinotermitidae). Biol. Control 30:523-529. [Google Scholar]
- 38.Watanabe, K., K. Abe, and M. Sato. 2000. Biological control of an insect pest by gut-colonizing Enterobacter cloacae transformed with ice nucleation gene. J. Appl. Microbiol. 88:90-97. [DOI] [PubMed] [Google Scholar]
- 39.Waterfield, N., A. Dowling, S. Sharma, P. J. Daborn, U. Potter, and R. H. ffrench-Constant. 2001. Oral toxicity of Photorhabdus luminescens W14 toxin complexes in Escherichia coli. Appl. Environ. Microbiol. 67:5017-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterfield, N., M. Hares, G. Yang, A. Dowling, and R. ffrench-Constant. 2005. Potentiation and cellular phenotypes of the insecticidal toxin complexes of Photorhabdus bacteria. Cell. Microbiol. 7:373-382. [DOI] [PubMed] [Google Scholar]
- 41.Wright, M. S., A. K. Raina, and A. R. Lax. 2005. A strain of the fungus Metarhizium anisopliae for controlling subterranean termites. J. Econ. Entomol. 98:1451-1458. [DOI] [PubMed] [Google Scholar]
- 42.Yanagawa, A., and S. Shimizu. 2007. Resistance of the termite, Coptotermes formosanus Shiraki, to Metarhizium anisopliae due to grooming. Biocontrol 52:75-85. [Google Scholar]
- 43.Yanagawa, A., F. Yokohari, and S. Shimizu. 2008. Defense mechanism of the termite, Coptotermes formosanus Shiraki, to entomopathogenic fungi. J. Invertebr. Pathol. 97:165-170. [DOI] [PubMed] [Google Scholar]
- 44.Yu, H., D. H. Gouge, and P. Baker. 2006. Parasitism of subterranean termites (Isoptera: Rhinotermitidae: Termitidae) by entomopathogenic nematodes (Rhabditida: Steinernematidae; Heterorhabditidae). J. Econ. Entomol. 99:1112-1119. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, X. J., F. Z. Sun, T. C. Zhao, A. Y. Ding, and C. R. Tang. 2004. A stably transgenic INA Enterobacter cloacae for control of insect pests. Sci. Agric. Sin. 37:227-232. [Google Scholar]
- 46.Zoberi, M. H. 1995. Metarhizium anisopliae, a fungal pathogen of Reticulitermes flavipes (Isoptera: Rhinotermitidae). Mycologia 87:354-359. [Google Scholar]