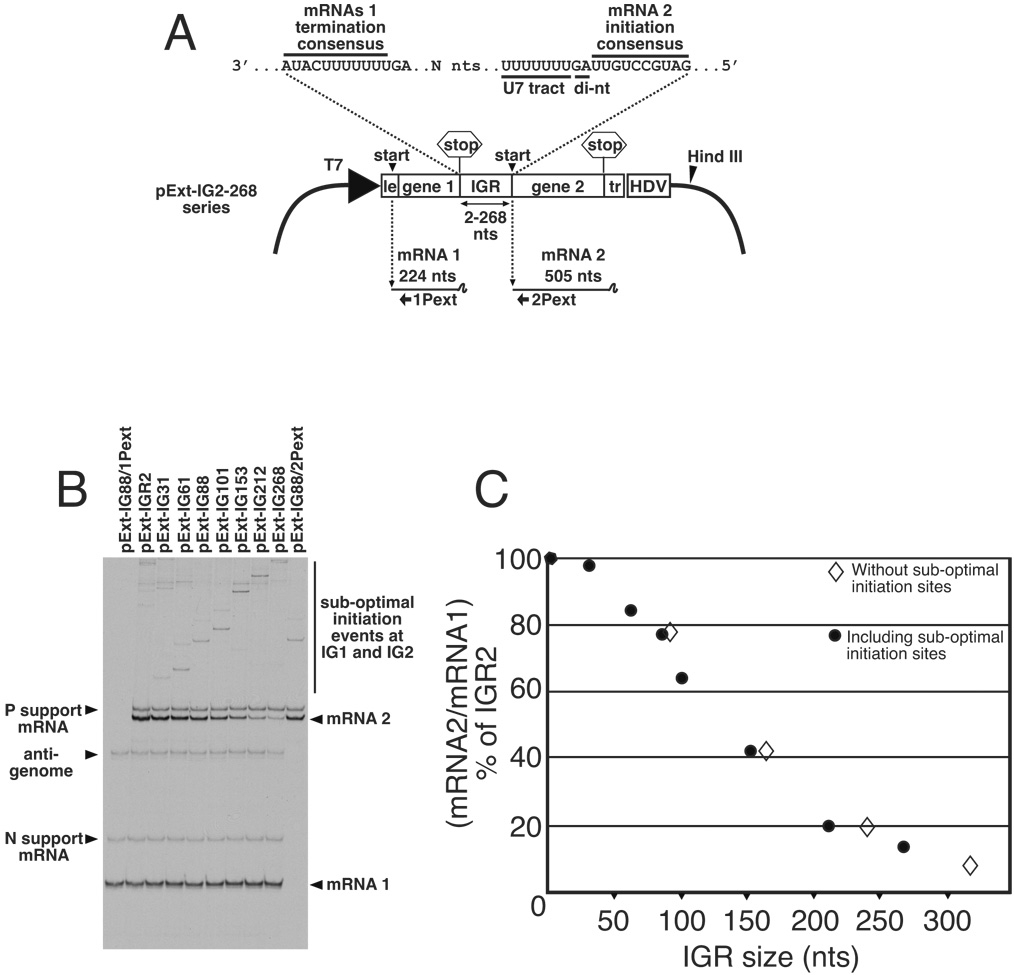
Figure 1. Analysis of the transcription activity of bicistronic genome analogs with lengthened intergenic regions (IGRs).
(A). Schematic representation of plasmid pExt-IG and its derivatives. These plasmids were individually transfected into vTF7-3 infected BHK cells along with support plasmids expressing VSV N, P and L ORFs, where they expressed an RNA representing a VSV anti-genomic analog. Encapsidation and replication of this RNA yielded a genome analog containing upstream (mRNA1) and downstream (mRNA2) transcriptional units separated by IGRs ranging between 2 and 268 nucleotides in length. Locations of U7 tract and intergenic di-nucleotide (di-nt) 3’ of the mRNA 2 initiation signal are shown. (B). Abundance of mRNA1 and mRNA2 transcribed from bicistronic templates was determined by dual extension of end-labeled oligonucleotides 1Pext and 2Pext, respectively. RNAs generated from template pExt-IG88 were also subjected to single primer extension reactions using individually 1Pext and 2Pext, to show position of mRNA 1 and mRNA2 respectively. Extension products were electrophoresed on a denaturing 6% acrylamide sequencing gel, and visualized by autoradiography. (C). The ability of the VSV RdRp to access the downstream gene start was calculated by quantitation of radiolabeled products representing downstream mRNA2 as a fraction of upstream mRNA1. This figure was then expressed as a percentage of that determined for template IGR2, which has the wild-type IGR length of 2 nts, and plotted as closed circles. Templates having the sub-optimal IGR start sites removed by mutagenesis were analyzed in the same way, and the values for mRNA2/mRNA1 % of IGR2 are also plotted as open diamonds.