Abstract
Merozoite surface proteins have been implicated in the initial attachment to the host red blood cell membrane that begins the process of invasion, an important step in the life cycle of the malaria parasite. In Plasmodium falciparum, merozoite surface proteins include several glycosylphosphatidyl inositol-anchored proteins and peripheral proteins attached to the membrane through protein-protein interactions. The most abundant of these proteins is the merozoite surface protein 1 (MSP1) complex, encoded by at least three genes: msp1, msp6, and msp7. The msp7 gene is part of a six-member multigene family in Plasmodium falciparum. We have disrupted msp7 in the Plasmodium falciparum D10 parasite, as confirmed by Southern hybridization. Immunoblot and indirect immunofluorescence analyses confirmed the MSP7 null phenotype of D10ΔMSP7 parasites. The synthesis, distribution, and processing of MSP1 were not affected in this parasite line. The level of expression and cellular distribution of the proteins MSP1, MSP3, MSP6, MSP9, and SERA5 remained comparable to those for the parental line. Furthermore, no significant change in the expression of MSP7-related proteins, except for that of MSRP5, was detected at the transcriptional level. The lack of MSP7 was not lethal at the asexual blood stage, but it did impair invasion of erythrocytes by merozoites to a significant degree. Despite this reduction in efficiency, D10ΔMSP7 parasites did not show any obvious preference for alternate pathways of invasion.
The pathological consequences of infection by the human malaria parasite, Plasmodium falciparum, are mainly due to its asexual life cycle within the bloodstream. Invasion of an erythrocyte by the merozoite form of the parasite initiates this process, followed by intracellular development and multiplication, leading to the release of new merozoites that invade further red blood cells. Electron microscopy studies reveal a thick fibrillar surface protein coat surrounding the merozoite, which is involved in the initial and probably reversible contact of the parasite with the host erythrocyte (3, 16). Proteins residing on the merozoite surface and within apical organelles have been implicated in specific interactions with host receptors to initiate invasion of erythrocytes. These proteins are also key targets of protective antibody responses, and as a consequence, several have emerged as promising vaccine candidates for control of P. falciparum infections. The surface coat of the merozoite is composed of a number of proteins, the most abundant of which is a complex of three merozoite surface proteins (MSPs): MSP1, MSP6, and MSP7 (5, 42, 46). Glycosylphosphatidyl inositol (GPI) membrane-anchored MSP1 is processed during maturation of merozoites and erythrocyte invasion (5-7, 22, 41). Following invasion, only a C-terminal 19-kDa fragment (MSP119) is retained on the parasite membrane within the newly invaded erythrocyte. The rest of the MSP1 protein is shed as a protein complex comprising four polypeptides of MSP1 (83, 38, 30 and 33 kDa) and associated 36- and 22-kDa polypeptides derived from MSP6 and MSP7, respectively (6, 41, 42).
MSP7 is synthesized as a 48-kDa precursor and binds full-length MSP1 in a pre-Golgi compartment (31). Prior to its translocation to the membrane, MSP7 undergoes a first proteolytic cleavage, releasing an N-terminal 20-kDa fragment (MSP720), while the C-terminal 33-kDa fragment (MSP733) remains in association with the MSP1 precursor (31). The second proteolytic processing of MSP733 yields a 22-kDa C-terminal fragment (MSP722) and occurs along with the primary processing of MSP1 (31). Upon erythrocyte invasion by the merozoite, MSP7 can be detected as a 22- or 19-kDa component of the shed MSP1 complex, depending upon the parasite line and the presence of Gln or Lys at position 194 in the MSP7 amino acid sequence. Apart from its association with MSP1 and sequential proteolytic processing coincident with merozoite development and maturation, nothing is known about the role of MSP7 in the biology of Plasmodium species. While an involvement in primary recognition and attachment to erythrocytes is likely for at least some of the surface proteins, including MSP7, to date there is little experimental evidence available. For example, a number of investigators have reported binding of MSP1 or MSP1 fragments to erythrocyte receptors (29), and more-recent data suggest that MSP119 binds to erythrocyte band 3. Similar interactions with band 3 have been reported for MSP7-derived peptides (17). While antibodies that target MSP7 can significantly inhibit invasion (23), they do not completely eliminate parasite entry into erythrocytes. Genetic redundancy or alternate pathways of invasion may explain such partial inhibition. However, as is the case with all other surface proteins, the biological significance of MSP7-receptor interactions has not been confirmed using reverse-genetics approaches.
The gene encoding MSP7 is located on chromosome 13 of P. falciparum. There are five downstream open reading frames (ORFs) at this locus with significant similarity to msp7 that form a multigene family (27). These ORFs, encoding MSP7-related proteins (MSRP1 to -5), are present in the transcriptome of trophozoite and schizont stages (21, 25), and it has been proposed that the msp7 gene family is an example of genetic redundancy (27), although their products were not detected in the merozoite proteome (15). To date, only MSRP1 (encoded by Pf13_0196) has been detected in the parasite, and that was in the detergent-resistant membrane fractions of P. falciparum 3D7 schizonts (39). In the syntenic chromosomal region of the rodent malaria parasites, Plasmodium yoelii and Plasmodium berghei, there are three orthologues to P. falciparum msp7 at the locus (27, 44, 46). In P. berghei, the gene designated msp7 is in the middle of the cluster and has been successfully disrupted, leading to a null phenotype without any lethal consequences to the parasite but switching the parasite into a more pronounced predominantly reticulocyte-tropic cell preference (44).
Molecular genetic approaches, especially the generation of gene knockout lines, have proved very useful in defining the relevance of interactions between parasite proteins and host cell receptors. For example, two distinct families of apical protein families—the Duffy binding-like proteins, referred to for P. falciparum as erythrocyte binding antigens (2, 18, 19), and the reticulocyte binding protein homologue family (35, 36)—have been targeted and disrupted successfully, resulting in altered invasion phenotypes. On the other hand, gene disruption technology has been somewhat less successful for the analysis of merozoite surface proteins, except for MSP5 (40) and MSP8 (4, 12), in which cases no discernible phenotypes were observed.
To characterize the role of MSP7 in the erythrocytic cycle of P. falciparum, in this study we successfully disrupted the msp7 gene in the D10 parasite line. The gene knockout line, D10ΔMSP7, did not produce detectable levels of MSP7, but the parasites were viable. The absence of MSP7 had no effect on the subcellular distribution of several late-stage antigens and no effect on the proteolytic processing of MSP1. Nevertheless, the absence of MSP7 significantly impaired erythrocyte invasion.
MATERIALS AND METHODS
Parasites.
P. falciparum (D10) and the msp7 knockout line were cultured in RPMI 1640 plus Albumax II using human O-positive blood cells. The parasites were synchronized using Percoll and sorbitol techniques (33). Merozoites were collected as described elsewhere (8, 43).
Nucleic acid techniques and transfection.
msp7 DNA fragments were generated by PCR amplification from P. falciparum (D10) genomic DNA using the oligonucleotide pairs ACGTCCGCGGCATTGTTTTTATTCGTATACAATTGGAAT and AGCTAGATCTCCTAAGAATGTGTCATCATTCATATCTAATTC for 5′ target sequences or ACGTCCATGGACAAAATAACGATTCACATTTTGAAAATGTTGATG and AGCTCCTAGGCCTATTAATACTTATATAAAGGTACACAATTTAACCGAATG for 3′ target sequences (restriction sites are underlined). These DNA fragments were cloned in the plasmid pHTK (13) at the SacII/BglII and NcoI/AvrII sites to obtain the plasmid pHTKΔMSP7, which was then used to transfect P. falciparum D10 ring-stage infected erythrocytes as described previously (14). The transfected parasites were selected on WR99210 and later with ganciclovir to obtain MSP7 knockout parasites by using standard procedures (13). Genomic DNA was isolated using DNAzol (Invitrogen), and total RNA was isolated from trophozoites and schizonts in culture using Trizol (Invitrogen). Hybridization analysis was carried out according to standard procedures; the DNA probes corresponding to MSRP ORFs (sequences of chromosome 13 corresponding to msrp1 were 1402487 to 1403629; for msrp2, 1397271 to 1400118; for msrp3, 1394929 to 1396116; for msrp4, 1389693 to 1390622; and for msrp5, 1387068 to 1388447; GenBank accession NC_004331) were PCR amplified from 3D7 genomic DNA and radiolabeled.
Immunoblotting and immunoprecipitation.
Percoll-purified schizont-stage parasites (40 to 48 h postinvasion) and merozoites were solubilized in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer with dithiothreitol as a reducing agent and then fractionated on 12% Novex Nu-PAGE gels. Proteins were electrophoretically transferred to Protran nitrocellulose (Schleicher & Schuell, Keene, NH), and immunoblotting was performed as described previously (31) with rat polyclonal antibodies against immunoglobulin binding protein (BiP) orthologue (obtained from MR4), rabbit polyclonal antibodies to the merozoite surface proteins MSP1 (6), MSP3 (28), and MSP7 (31), or mouse polyclonal antibodies against MSP6 (46). Bound antibody was detected with either horseradish peroxidase-coupled goat antimouse or antirabbit antibody (Bio-Rad Laboratories) and developed by enhanced chemiluminescence (GE Healthcare), according to the manufacturer's instructions.
Percoll-enriched late-stage parasites were radiolabeled with Promix (GE healthcare) for 2 h and solubilized in NP-40- or SDS-containing buffers, and the extracts used in immunoprecipitation studies with mouse monoclonal antibody (MAb) to MSP119, MAb 12.8 (8), and rabbit polyclonal antibodies to MSP1 or MSP7 as described earlier (31). For visualizing of MSP1 processing, radiolabeled parasites were allowed to develop for a further 6 h prior to solubilization.
Indirect immunofluorescence analysis.
Synchronized late-schizont-stage parasites were smeared, air dried, and fixed in 1% formaldehyde prepared fresh in phosphate-buffered saline for 5 min at room temperature and then immediately permeabilized with dry acetone at −20°C for 5 to 10 s. The smears were then incubated with 0.2% fish skin gelatin (Sigma), followed by a combination of antibodies against P. falciparum proteins and Alexa Fluor-labeled goat antimouse and antirabbit antibodies (Molecular Probes). Rabbit polyclonal antibodies against MSP9 (24) and SERA5 (30) were obtained from Vir Chauhan and Toshihiro Horii, respectively. The DNA stain, Hoechst 33428, was included in the second antibody incubation step. Dual-color fluorescence images were captured using a digital camera and fluorescence microscope (Zeiss, Gottingen, Germany).
Red blood cell treatment and parasite growth and invasion assay.
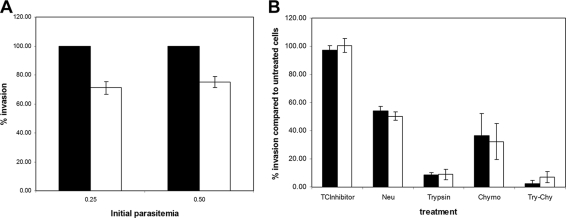
To determine the growth and invasion rate of the parasite lines D10 and D10ΔMSP7, cultures of synchronized trophozoites grown in the absence of WR99210 (27 to 30 h postinvasion) were diluted with fresh erythrocytes to an approximate initial parasitemia of 0.2 to 0.5%. Parasites were then incubated in culture for 48 h and stained with 0.1 mg/ml ethidium bromide, and the final parasitemia level was determined by using a FACScalibur flow cytometer (Becton Dickinson) as described previously (34). The growth and invasion rate was expressed as the percentage of invasion compared to that of the control parental line. The confidence interval (CI) (95%) was determined from triplicate replicates of each condition. The significance of difference between the D10 and D10ΔMSP7 lines was determined by the one-way analysis of variance (ANOVA).
Digestion of erythrocyte surface molecules and invasion assays were performed essentially as described previously (11) with the following modifications. Cultures of synchronized trophozoites of D10 and D10ΔMSP7 parasites were adjusted to 1% parasitemia and 2% hematocrit with fresh red blood cells. Aliquots of 0.9 ml were treated with buffered medium (control) or buffered medium containing either Vibrio cholerae neuraminidase (0.10 U/ml; Calbiochem), l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (1.2 mg/ml; Sigma), or α-chymotrypsin (1.2 mg/ml; Sigma) at 37°C for 75 min and then washed. Following treatment, protease-treated parasite-infected red blood cells were incubated with trypsin-chymotrypsin inhibitor (Sigma) at a final concentration of 0.5 mg/ml for 10 min at ambient temperature and then washed three times. The treated parasites were resuspended in culture medium at a hematocrit value of 2%, and three 150-μl aliquots were dispensed in a 96-well culture plate. Following invasion, each well of a triplicate set was analyzed individually by counting 100,000 red blood cells. Red blood cells infected with reinvaded parasites were identified based on fluorescence, and the fluorescence-activated cell sorting data were analyzed using the BD CellQuest Pro V5.2.1 software program. The invasion rates were normalized with reference to control buffer-treated culture, and statistical significance was determined as described above.
Growth and invasion assay in mixed culture.
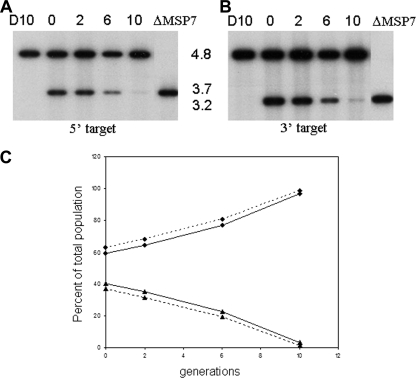
Cultures of P. falciparum D10 or D10ΔMSP7 trophozoite-infected red blood cells were mixed, and the mixed culture was maintained at about 2% parasitemia and 1% hematocrit by adding fresh red blood cells for a period corresponding to 20 generations. Genomic DNA was isolated from these parasites after 0 (about 6 h after mixing), 2, 6, 8, and 10 generations by using DNAzol, and Southern hybridization was performed using standard procedures. The autoradiograph was scanned and analyzed by the ImageJ software program (1) to quantify signal intensities, which were then converted into percent parasite population.
RESULTS
Disruption of the P. falciparum msp7 gene.
To determine whether MSP7 performs a function essential to the erythrocytic cycle, we attempted to disrupt the msp7 gene by using a thymidine kinase-based negative selection system (13). Two pairs of gene-specific primers were used to amplify MSP7 DNA fragments, each including 5′ or 3′ parts of MSP7 ORF and noncoding regions (PLASMODB [www.plasmodb.org] chromosome 13 nucleotides 658542 to 657686 for the 5′ target and 657598 to 656662 for the 3′ target). These fragments were then cloned into pHTK on either side of the human dihydrofolate reductase (hDHFR) gene to obtain the knockout construct pHTKΔMSP7. P. falciparum D10 asexual-stage parasites were transfected by electroporation using the knockout construct (Fig. 1A). Transfected parasites with episomal plasmid or integrated DNA, selected with WR99210, were first observed by days 16 to 20 postelectroporation. The mixed drug-resistant population of parasites was then subjected to three alternating cycles of growth with and without WR99210 to eliminate parasites containing only episomal plasmid DNA. After two or three such cycles, the cultures were treated with ganciclovir to select for parasites with DNA integrated by double-crossover recombination and to eliminate parasites carrying episomes or DNA integrated due to single crossovers at 5′ or 3′ target regions. Such double recombination at the MSP7 locus was predicted to result in a knockout of the MSP7 gene in parasites, which were subsequently cloned by limiting dilution.
FIG. 1.
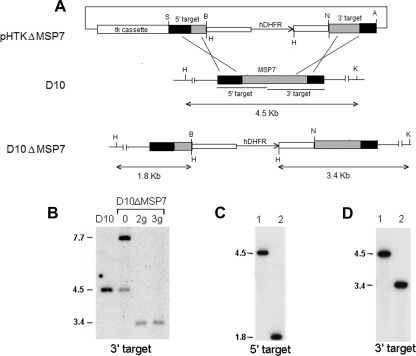
Disruption of Plasmodium falciparum D10 msp7 gene. (A) Schematic representation of the knockout plasmid construct (pHTKΔMSP7), the msp7 locus of the parental parasite line (D10), and the resultant msp7 knockout parasite (D10ΔMSP7) lines. The plasmid construct carries a viral thymidine kinase gene cassette (tk cassette) for negative selection with ganciclovir and an hDHFR cassette to insert into the D10 msp7 locus. Insertion of the hDHFR cassette by double homologous recombination within the 5′ and 3′ target regions of msp7 changes the hybridizing KpnI-HindIII restriction fragment sizes from 4.5 kb to 1.8 (with the 5′ target probe) or 3.4 kb (with the 3′ target probe). Relevant restriction sites—AvrII (A), BamHI (B), HindIII (H), KpnI (K), NcoI (N), and SacII (S)—are shown. (B) Southern hybridization of genomic DNA from D10 and transfected parasites digested with KpnI and HindIII and probed with msp7 3′ target probe. Lanes show transfected parasites (0) and ganciclovir-selected parasites after second (2g) or third (3g) WR22910 cycling. The signal at 7.7 kb in lane 0 corresponds to episomal DNA. (C and D) Hybridization analysis of genomic DNA from D10 (lane 1) or the D10ΔMSP7 clone (lane 2) with 5′ or 3′ target DNA probes, respectively. Molecular sizes of hybridizing DNA fragments are indicated.
The recombination of the knockout plasmid construct with the genomic locus, resulting in the disruption of the resident msp7 gene, is represented schematically in Fig. 1A. To confirm the integration of the DNA into the msp7 gene, we carried out Southern blot analysis of genomic DNA isolated from D10 and D10ΔMSP7 parasites with two MSP7-specific probes: the 5′ and 3′ targeting DNA fragments. As seen in Fig. 1B, only the disrupted msp7 locus was present following second (Fig. 1B, lane 2g) or third (Fig. 1B, lane 3g) ganciclovir selection of the transfected parasites. The resident MSP7 locus, detected as a 4.5-kb fragment in D10 DNA and the 9.1-kb episomal DNA present in lane 0, disappeared upon ganciclovir selection. The banding pattern was consistent with integration of a single hDHFR copy, leading to disruption of the targeted msp7 gene. A single clone, D10ΔMSP7, carrying an hDHFR cassette at the MSP7 gene, was then characterized. Figures 1C and D show the hybridization results of D10ΔMSP7 genomic DNA probed with 5′ or 3′ targets, revealing the absence of the resident intact msp7 gene and the presence of the altered locus with a single copy of the hDHFR cassette.
MSP7 protein is not detected in D10ΔMSP7 parasites.
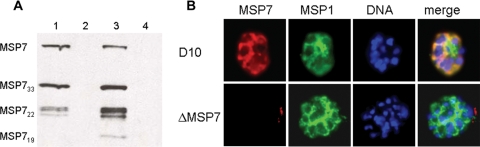
The expression of MSP7 in the D10 and D10ΔMSP7 parasite lines was examined in synchronized populations. MSP7 expression peaks in late-stage parasites, starting 30 h after invasion. Therefore, late-stage parasites were collected and protein extracts analyzed by Western blotting using anti-MSP7 antibodies (Fig. 2A). When extracts were probed with rabbit antisera directed to the C terminus of MSP7, the expected 45-kDa precursor and processed forms (33, 22 and 19 kDa) of MSP7 were observed only in the parental D10 parasite line (Fig. 2A, lanes 1 and 3). These signals were absent in the extract of the D10ΔMSP7 parasites (lane 2), thus verifying the MSP7-null phenotype expected from the gene disruption. Further, removal of drug selection from the culture of D10ΔMSP7 had no effect (lane 4), and the disrupted MSP7 locus was stably propagated in this clonal parasite line. These results were confirmed by immunoblotting extracts of parasites after 10 weeks of culture in the absence of WR99210. These results showed that disruption of the msp7 gene resulted in ablation of full-length MSP7 expression in the transfected lines. However, we cannot rule out the expression of a truncated N-terminal polypeptide in these parasites, which would be less than 100 amino acids in length.
FIG. 2.
MSP7 null phenotype of D10ΔMSP7 parasite clone. (A) Western blot analysis of late-stage D10 (lanes 1 and 3) or D10ΔMSP7 clone (lanes 2 and 4) with rabbit polyclonal antibodies to the MSP7 protein. The knockout parasite clone was cultured in the presence (lane 2) or absence (lane 4) of WR99210. Different forms of MSP7, such as the precursor (MSP7) and processed forms (MSP733, MSP722, and MSP719), are indicated. (B) Indirect immunofluorescence analysis of formaldehyde-fixed, acetone-permeabilized late-stage parasites from D10 and the D10ΔMSP7 clone with monoclonal anti-MSP1 (MSP1) and anti-MSP7 (MSP7) antibodies. DNA was visualized with Hoechst 33428. Colocalization was confirmed by color-merging (merge).
It has previously been shown that the localization of MSP7 by indirect immunofluorescence microscopy is consistent with a merozoite surface location in the 3D7 and T9/96 parasite lines (31, 32). Here the surface location of MSP7 in the D10 parasite line was confirmed by colocalization with MSP1 using indirect immunofluorescence with a rabbit polyclonal MSP7-specific antibody (Fig. 2B). In schizont-stage D10 parasites, an MSP1-specific antibody (MAb 12.8) stained the circumference of individual merozoites within the maturing parasite, consistent with the known location of MSP1 on the merozoite surface; MSP7 showed a similar staining pattern in the parental D10 line, consistent with its association with MSP1. Using this same antibody, MSP7 was not detected above background labeling in the D10ΔMSP7 parasites, further confirming the absence of the MSP7 protein. MAb 12.8 gave similar reactivities with the D10ΔMSP7 parasite and with the D10 parasite, indicating that the loss of MSP7 expression does not alter the trafficking and localization of MSP1 (Fig. 2B).
Absence of MSP7 does not affect MSP1 processing.
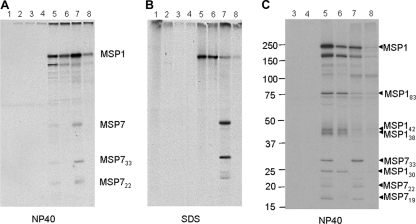
We have previously reported that the association of MSP7 with MSP1 is resistant to the nonionic detergent NP-40 but not to SDS and that the complex could be immunoprecipitated using either anti-MSP1 or anti-MSP7 antibody (32). Immunoprecipitation studies were undertaken to analyze the MSP1 complex and MSP1 and MSP7 interactions in both the D10 and D10ΔMSP7 lines, and the results are presented in Fig. 3. Anti-MSP1 antibodies (MAb 12.8) precipitated the MSP1 complex containing MSP7 and its fragments from extracts of radiolabeled D10 schizonts solubilized in NP-40-containing buffer (Fig. 3A, lane 5). Only MSP1 was precipitated with the same antibody from the D10ΔMSP7 parasite extract (Fig. 3A, lane 6). Likewise, the anti-MSP7 antibodies precipitated the complex containing the MSP7 fragments and MSP1 from NP-40-solubilized D10 parasites, but no MSP7 could be detected in the D10ΔMSP7 parasite extract (Fig. 3A, lanes 7 and 8). The detection of MSP7 or MSP1 in SDS-solubilized parasite extracts complemented the observations with NP-40-solubilized extracts (Fig. 3B). Small amounts of MSP1 were apparently immunoprecipitated with the anti-MSP7 antibodies from both NP-40- and SDS-solubilized parasites, possibly due to the presence of cross-reactive antibodies (Fig. 3A, lane 8, and B, lanes 7 and 8). Nonimmune mouse or rabbit serum did not precipitate any specific proteins (Fig. 3A and B, lanes 1 to 4).
FIG. 3.
Immunoprecipitation analysis of D10 and D10ΔMSP7 35S-biosynthetically labeled parasite extracts. (A and B) NP-40- or SDS-solubilized parasites from D10 (odd-numbered lanes) and D10ΔMSP7 clone (even-numbered lanes) immunoprecipitated with nonimmune mouse (lanes 1 and 2) or rabbit antisera (lanes 3 and 4), MSP1-specific MAb 12.8 (lanes 5 and 6), or rabbit polyclonal antibodies to MSP7 (lanes 7 and 8). (C) MSP1 processing in D10 and D10ΔMSP7 parasites allowed to develop for 6 h after radiolabeling. The sizes of molecular markers are shown in kDa. The figure labels are the same as those in panels A and B, except that polyclonal rabbit antibodies to MSP1 were used (lanes 5 and 6). The MSP1 precursor and its processed fragments (MSP183, MSP142, MSP138, and MSP130) and the precursor MSP7 and its processed forms (MSP733, MSP722, and MSP719) are indicated. The band below MSP1 at about 160 kDa in NP-40 extracts results from detergent-promoted cleavage of the MSP1 precursor.
Under the experimental conditions used above, the Percoll-enriched D10 or D10ΔMSP7 parasites had very little of the radiolabeled and processed MSP1 polypeptides or MSP722, as seen in the immunoprecipitations from both NP-40- and SDS-solubilized extracts (Fig. 3A and B). Therefore, radiolabeled parasites were allowed to develop for a further 6 h before harvesting and preparation of extracts in buffer containing NP-40. From these parasite lysates, rabbit polyclonal anti-MSP1 antibodies immunoprecipitated the MSP1 precursor as well as the 83-, 42-, 38-, and 30-kDa fragments. The results (Fig. 3C, lanes 5 and 6) were identical for the D10 and D10ΔMSP7 parasite extracts except for the absence of MSP7-specific bands in the complex from the knockout line. Anti-MSP7 antibodies immunoprecipitated the same complex from the D10 parasite extract (Fig. 3C, lane 7), but in contrast, they did not precipitate any MSP7 or fragments derived from it in the extract of the D10ΔMSP7 parasite (Fig. 3C, lane 8). Thus, the detection of processed forms of MSP1 in both parasite lines rules out any role for MSP7 in the primary proteolytic processing of MSP1.
A close examination of the MSP1 complex precipitated with anti-MSP1 antibodies from the NP-40 extracts of the D10ΔMSP7 parasite failed to reveal any new polypeptides that had taken the place of MSP7 in the MSP1 complex (Fig. 3A and C, lanes 6).
Absence of MSP7 does not alter expression and distribution of several other peripheral merozoite surface proteins.
In addition to MSP7, several other proteins have been reported to interact with the MSP1 complex through protein-protein interactions. We investigated expression and distribution of MSP3, MSP6, and MSP9 (ABRA) and a parasitophorous vacuolar protein, SERA-5, in order to examine the consequence of altering the nature of the MSP1 complex. The expression of the MSRPs was also analyzed at the transcriptional level.
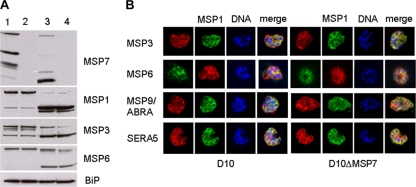
Immunoblot analysis of parasite extracts from D10 and the corresponding MSP7 knockout line with polyclonal antisera specific to MSP1, MSP3, or MSP6 did not reveal any difference in the amount of the protein between the two lines (Fig. 4A) in either late-stage intracellular parasites (lanes 1 and 2) or free merozoites (lanes 3 and 4). The results were identical whether or not the selection drug, WR99210, was present during the culture of the knockout parasite line. A corresponding membrane probed with antibodies against the immunoglobulin binding protein (BiP) was used as a control to monitor the total amount of protein loaded from each parasite population; the results confirmed that for each sample, approximately equal amounts of total protein from each parasite population were loaded (Fig. 4A).
FIG. 4.
Analysis of the expression of peripheral merozoite surface and vacuolar proteins by immunoblotting and indirect immunofluorescence. (A) Immunoblot analysis of late-stage parasites (lanes 1 and 2) and purified merozoites (lanes 3 and 4) from D10 (lanes 1 and 3) or D10ΔMSP7 (lanes 2 and 4) cloned parasites with antisera to the MSP7, MSP1, MSP3, and MSP6 proteins. Antibodies to immunoglobulin binding protein (BiP) orthologue were used to ascertain that equal amounts of proteins were loaded. (B) Indirect immunofluorescence analysis of D10 and D10ΔMSP7 parasites. Fixed smears of late-stage parasites were probed with antibodies to MSP3, MSP6, MSP9, and SERA5 together with anti-MSP1 antibodies. Rabbit polyclonal antibodies are in red, while mouse antibodies are in green. Hoechst 33428 was used to visualize DNA (blue). Color-merging was carried out to analyze colocalization with MSP1 (merge).
To examine the effect of MSP7 disruption on the subcellular distribution of MSP3, MSP6, MSP9/ABRA, and SERA5, we compared the MSP7 knockout with the parental D10 line by indirect immunofluorescence using antiserum specific to each protein in conjunction with anti-MSP1 antibodies (Fig. 4B) and schizont-stage parasites, since MSP7 may play a role in the development of the merozoites and their release, or invasion. All the antibodies, however, gave similar patterns against both the D10 and D10ΔMSP7 parasites. Whereas the antibodies to MSP3, MSP6, and SERA-5 revealed a clear colocalization with MSP1, the locations of MSP9/ABRA and MSP1 show some overlap but also some distinct areas for both parasite lines.
The effect of knocking out the msp7 gene on the transcription of the genes for the MSP7-related proteins was examined to determine whether or not they might be upregulated as a compensatory mechanism. mRNA was prepared from both D10 and D10ΔMSP7 parasites and analyzed by Northern blotting using gene-specific probes (Fig. 5). msp7 mRNA was clearly absent in the knockout parasites. For most of the MSRP genes, the level of transcripts was approximately the same in both mRNA preparations, although the msrp1 transcript was poorly detected. The only exception was msrp5 (177.4% ± 28.3%, mean percentage ± 95% CI compared to D10 msrp5), for which a near-twofold increase in the transcript level was observed. The significance of this increase (P < 0.01) is not clear since there exists no evidence of the MSRP5 protein in the parasites in spite of the transcript.
FIG. 5.
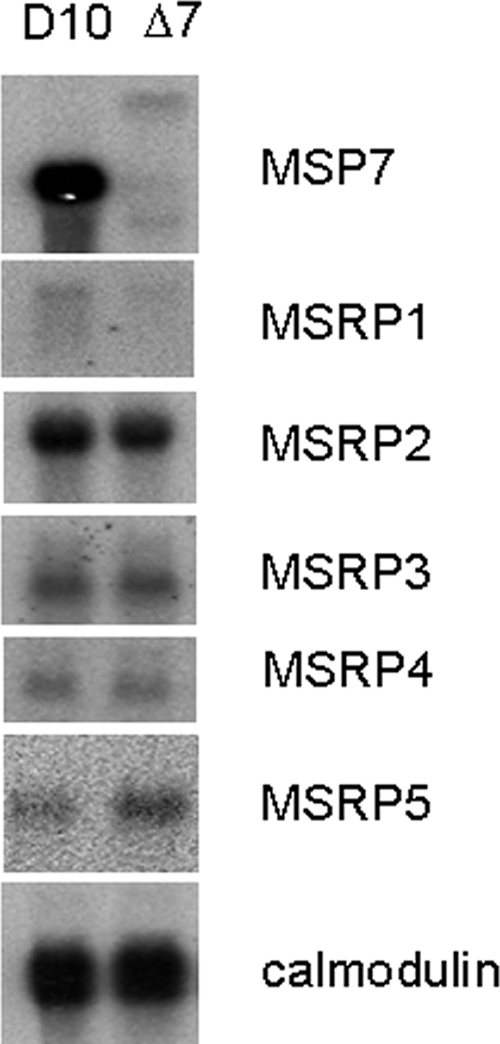
Transcription at msp7 locus. Northern blot analysis of late-stage (33 to 36 h) parasites from D10 and D10ΔMSP7 clones using DNA probes specific to MSP7 and MSP7-related protein genes (MSRP1 to MSRP5). A calmodulin cDNA probe was used to confirm equal loading of RNA in each lane.
Disruption of MSP7 reduces merozoite invasion of red blood cells.
To determine whether disruption of MSP7 had any effect on the ability of merozoites to invade red blood cells, the D10 and D10ΔMSP7 clones were compared in merozoite invasion assays conducted on a number of occasions in triplicate (0.2 and 0.5% starting parasitemias are shown in Fig. 6A). The results from three independent experiments were analyzed to determine CIs, and the significance was examined by one-way ANOVA. A significant difference (P < 0.001) was observed in the invasion rates between the parental D10 and the D10ΔMSP7 lines; the knockout line consistently displayed about a 29% ± 9.1% (mean percentage ± 95% CI) reduction in erythrocyte invasion. This difference was not due to fewer merozoites being produced in the schizonts of the MSP7 knockout line. The number of merozoites formed in each mature schizont prior to rupture was counted from Giemsa-stained smears, and a similar distribution (median, 18.0; n = 75) was observed (D10, 17.88 ± 0.22; D10ΔMSP7, 17.95 ± 0.10).
FIG. 6.
Effect of msp7 gene disruption on erythrocyte invasion by merozoites. (A) Invasion assay using O-positive erythrocytes and an initial parasitemia of 0.25 or 0.5%. The level of invasion was determined by flow cytometry and then normalized to that of the parental D10 parasite line. Values are represented as percent invasion compared to that of D10 parasites, with 95% CIs calculated from three independent assays as error bars. (B) Relative levels of invasion into enzyme-treated erythrocytes for the parental D10 line (▪) and the D10ΔMSP7 (□) clone. The erythrocytes in trophozoite cultures (1% parasitemia) were treated with neuraminidase (Neu), trypsin, chymotrypsin (Chymo), or trypsin and chymotrypsin (Try-Chy), and then the parasitemia was determined after 48 h. In control experiments, the cells were treated with buffer alone. An appropriate control for trypsin-chymotrypsin inhibitor treatment alone (TCInhibitor) was included. The levels of invasion were normalized to that in buffer-treated control cultures to yield percent invasion. The results are the means of three independent assays, and the error bars indicate 95% CIs.
In order to examine whether the loss of MSP7 expression led to a switch in the erythrocyte invasion pathway to one distinct from that of the D10 parental parasite, the invasion of erythrocytes treated with enzymes known to cleave erythrocyte receptors involved in parasite invasion was compared. The results from three independent experiments were analyzed to determine CIs, and the significance was examined by one-way ANOVA. No significant difference (P > 0.05) in the rate of invasion of erythrocytes treated with trypsin, neuraminidase, chymotrypsin, or chymotrypsin plus trypsin was observed between the D10ΔMSP7 and D10 wild-type lines (Fig. 6B). The D10 line seems to use a trypsin-sensitive pathway, in addition to a sialic-acid-dependent pathway (45). This preference appears unaltered by the loss of MSP7 but differs from the previously reported 3D7 parasite line use of a trypsin-sensitive and neuraminidase- and chymotrypsin-insensitive invasion pathway (45).
MSP7 null parasites have a growth disadvantage in a mixed population with the parental line.
Having confirmed the deletion of msp7 in D10ΔMSP7 parasites and examined its effect on erythrocyte invasion in a single-cycle assay, we investigated growth over 10 generations in a mixed population of D10ΔMSP7 and D10 parasites. Starting with approximately equal numbers of the two parasites in a mixed culture (D10:D10ΔMSP7, 60:40), the culture was maintained over multiple generations, diluting the parasitemia every 48 h to less than 2%. At specific time points, genomic DNA was prepared from the parasite, restricted with EcoRI, and subjected to Southern blotting with msp7-specific probes (Fig. 7). Comparison of DNA signal intensities of the bands corresponding to the wild type (4.8 kb) and the disrupted msp7 loci (3.2 or 3.7 kb) showed that under these mixed-culture conditions, D10ΔMSP7 parasites were disadvantaged relative to the wild-type parasite, and by the 10th generation, they were hardly detectable, as seen by a reduction of the signal at 3.2 or 3.7 kb. The signal intensities were quantified from the scanned image, converted to percentages of the total parasite population for each time point, and plotted against the generation (Fig. 7C). A steady decline in the knockout parasite line and a concomitant increase in parental parasites were observed in the mixed culture, further supporting the invasion defect observed in a single-cycle assay.
FIG. 7.
Growth assay in a mixed culture of D10 and D10ΔMSP7 cloned parasites determined by DNA hybridization analysis. (A and B) Genomic DNA isolated from D10, the D10ΔMSP7 (ΔMSP7) clone, and a mixed culture of the two after 0, 2, 4, 6, or 10 generations was restricted with EcoRI and probed with DNA corresponding to the 5′ (A) or 3′ (B) target sequence, as explained in the legend to Fig. 1. Molecular sizes of msp7-hybridizing restriction fragments of D10 (4.8 kb) and D10ΔMSP7 (3.7 or 3.2 kb) are indicated. (C) Signal intensities determined by using ImageJ from each lane of panel A (solid line) or panel B (dashed line) were plotted as percent parasite population versus number of generations. ⧫, D10; ▴, D10ΔMSP7.
DISCUSSION
Previous data suggested that MSP7 is an important molecule on the merozoite surface. MSP7 forms a tight complex with MSP1 during biosynthesis, and both molecules are processed by specific protease cleavages (31). On the merozoite surface, a 22-kDa fragment of MSP7 is an integral component of the MSP1 complex and was estimated to be present in amounts stochiometrically equal to those of the MSP1-derived polypeptides. The protein is abundant on the parasite surface, as judged by radioiodination of intact merozoites, and the target of antibodies that inhibit erythrocyte invasion. At the sequence level, the protein is highly conserved across different lines of P. falciparum, and orthologous genes have been detected in all other Plasmodium species examined. Despite these facts, we show here that the MSP7 gene can be deleted without severely impairing the parasite's asexual erythrocytic life cycle and in contrast to attempts to delete the MSP1 gene, which have proved to be futile, presumably because the protein serves an essential function. Successful disruption of the msp7 gene in the parasite genome indicates that this gene is not essential for parasite survival.
The consequence of deleting the msp7 gene was difficult to detect. There was no apparent effect on the biosynthesis and trafficking of MSP1 to the merozoite surface or the proteolytic processing of this molecule. The synthesis and localization of other molecules that have been proposed to interact with MSP1 or other proteins on the merozoite surface, including MSP6, MSP3, and MSP9/ABRA, also appeared to proceed normally in the D10ΔMSP7 parasite. There was no apparent affect on the level of transcription of the genes for most of the other members of the msp7 family. The ability of the parasite to invade red cells that had been treated with different enzymes was identical to that of the D10 parent, suggesting that there had been no switch to the use of alternate invasion pathways.
Despite our inability to detect any consequence of msp7 deletion at the molecular level, a reproducible phenotype was identified: a reduction of 20% in the ability of the parasite to invade red blood cells. The consequences of this reduction were highlighted in experiments where the parental and MSP7 knockout lines were cultured together as a single population; within about 10 generations, the wild-type parasites had outgrown the parasites with the deleted MSP7 gene. This result highlights the fact that relatively small differences in fitness can have profound effects on a parasite's ability to survive. Small phenotypic differences, for example, a few percent differences in invasion efficiency, which may result from host and parasite heterogeneity, epigenetic switching on or off of specific genes, or the consequences of reverse-genetics studies, may be difficult to measure experimentally but are likely to be important in parasite-parasite competition and parasite interaction with the host. Although in the current study the observed phenotype was detrimental, deletion of msp7 may be advantageous under different growth conditions; for example, it is possible that this deletion may improve the ability of the parasite to invade younger cells, as suggested by the results of deleting the second gene in the MSP7 locus of P. berghei (44), in which a preference for reticulocytes was increased. The in vitro culture conditions used in the current study would not detect an increased ability of the parasite to invade reticulocytes. It has been shown that antibodies to MSP7 inhibit parasite growth in vitro (23). These data are consistent with the phenotype of the parasite with the msp7 deletion characterized here.
In all Plasmodium genomes examined so far, msp7 is a member of a small family of related genes located at the same locus. For example, P. falciparum has MSP7 and five MSP7-related genes, and in rodent parasites, there are three genes in the family. The degree of sequence similarity between both the paralogous and orthologous proteins is relatively low but is greatest toward the C terminus. It is thought that the msp7 gene family arose due to a genetic amplification event given their similar organization, structural features, and adjacent chromosomal location (27). There is evidence that some or all of these genes are expressed (this study and others, including proteome analyses), and therefore, it is possible that one or more MSRPs may substitute for MSP7. Such an interpretation would be supported by results of pull-down experiments (27) suggesting that other members of the family can bind to MSP1 in addition to MSP7. However, in the experiments reported here, there was no evidence of MSRPs bound to the MSP1 complex from the D10ΔMSP7 parasite. It is possible that if the affinity of an MSRP for MSP1 is less than that of MSP7, then the interaction may not be stable in the presence of the NP-40 detergent. However, other members of the family have not been detected in the merozoite proteome or in the MSP1 complex, and we found no evidence of increased transcription. Recent studies of late-stage schizonts reported the abundant presence of MSP1 and MSP7 in the detergent-resistant membrane-fraction proteome (38, 39). Of the five MSP7-related proteins, only MSRP1 (PF13_0196) was found in this proteome (39). This is unlike the rodent parasite P. yoelii, in which all the three gene products could be detected (27; M. Kadekoppala and A. A. Holder, unpublished data). It will be of interest to determine whether or not the entire MSP7 locus can be deleted from the parasite genome.
In addition to the GPI-anchored proteins (MSP1, -2, -4, -5, -8, and -10), several peripheral proteins, such as MSP3, MSP6, MSP7, and MSP9/ABRA, have been suggested to be merozoite surface proteins. Two of the peripheral proteins, MSP6 and MSP7, were found in the MSP1 complex (41). The presence of MSP6 in the shed MSP1 complex has been unequivocally established (46). However, all of these proteins with the exception of MSP7 are predominantly soluble and in the parasitophorous vacuole; the absence of MSP3, MSP6, and MSP9/ABRA from the detergent-resistant membrane fraction proteome is consistent with this observation. Although MSP3 and MSP9 are frequently referred to as surface proteins, their relationship with the merozoite membrane or a membrane-anchored protein partner is yet to be defined. MSP7 has been reported to lack significant sequence variation between P. falciparum isolates from various geographical locations (31, 32), suggesting that it is not subject to strong immune selection. Attempts to disrupt many of the GPI-anchored surface proteins, including MSP1, have failed; however, MSP5 and MSP8 were disrupted without any apparent phenotype (4, 12, 40).
The enzyme treatments of red blood cells employed in this study are commonly used as a tool to characterize Plasmodium merozoite invasion pathways: sialic acid-dependent/trypsin-sensitive (associated with glycophorin A), sialic acid-dependent/trypsin-insensitive (glycophorin B), and sialic acid-independent/trypsin-sensitive (receptor X) (10). Using these defined invasion pathways, the D10ΔMSP7 clone did not exhibit an erythrocyte invasion phenotype different from that of its parental clone D10. Erythrocyte invasion by both the D10 and D10ΔMSP7 clones was reduced marginally but equally by treatment with neuraminidase, indicating that invasion is partially dependent on sialic acid residues on the erythrocyte surface. However, invasion of both lines was abolished when erythrocytes were treated with trypsin. Thus, parasite invasion assays with enzymatically treated erythrocytes, a process designed to expose a change in invasion pathways, failed to reveal any change in the invasion profile attributable to the loss of MSP7.
There have been several reports of the merozoite surface proteins interacting with red blood cell membrane proteins. Peptides derived from MSP1 and recombinant MSP119 appear to interact with band 3 (20). Similarly, several peptides, called high-activity binding peptides, derived from MSP3, MSP6, MSP7, and MSP9, could bind to the erythrocyte surface (9, 17, 26, 37). In MSP7, two regions in the N-terminal domain and the C-terminal 51-amino-acid residues have been proposed to bind erythrocytes (17); the significance of these findings remains unclear, since only the C-terminal region of MSP7 remains associated with MSP1 and the N-terminal region appears to be degraded (31, 32). The high-activity binding peptides corresponding to the C-terminal end of MSP7 might have a role in merozoite attachment to and invasion of red blood cells by interaction with band 3 (17). Results presented here show that the MSP7-null parasites do not display any differential effect with respect to the parental line in invasion assays using enzyme-treated red blood cells. This indicates that MSP7-band 3 interactions may not play a detectable significant role in merozoite invasion of erythrocytes.
It was suggested that sequential proteolytic cleavage of MSP7 and MSP1 may have a role in merozoite development and maturation (31). In particular, the first proteolytic cleavage of MSP7 occurs in a pre-Golgi compartment after its association with MSP1 precursor. In contrast, MSP1 primary processing occurs much later upon the surface of the developing merozoite, at the time of merozoite release. Our work shows that the presence of MSP7 and its consequent proteolytic processing are not prerequisites to MSP1 processing or merozoite development and release. We report here that the absence of MSP7 does not have any major consequences for MSP1 processing or parasite development and growth, with an only marginal, but significant, reduction in invasion of erythrocytes. Nevertheless, its conservation among all the Plasmodium species tested, in addition to its location on the merozoite surface, may suggest an importance of MSP7 in natural infections not revealed in the current study.
Acknowledgments
We thank Alan F. Cowman (for anti-MSP3 antibodies), Toshihiro Horii (for anti-SERA5 antibodies), V. S. Chauhan (for anti-MSP9/ABRA antibodies), and Judith Green (for calmodulin cDNA and MR4 [for anti-BiP antibodies]). We also thank Ellen Knuepfer and other members of the Holder lab for helpful discussions.
This work was supported in part by the UK Medical Research Council and the U.S. National Institutes of Health (HL078826).
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Abramoff, M. D., P. J. Magalhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 1136-42. [Google Scholar]
- 2.Adams, J. H., B. K. Sim, S. A. Dolan, X. Fang, D. C. Kaslow, and L. H. Miller. 1992. A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. USA 897085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, L. H., G. H. Mitchell, G. A. Butcher, E. D. Dennis, and S. Cohen. 1986. Structure and development of the surface coat of erythrocytic merozoites of Plasmodium knowlesi. Cell Tissue Res. 245281-290. [DOI] [PubMed] [Google Scholar]
- 4.Black, C. G., T. Wu, L. Wang, A. E. Topolska, and R. L. Coppel. 2005. MSP8 is a non-essential merozoite surface protein in Plasmodium falciparum. Mol. Biochem. Parasitol. 14427-35. [DOI] [PubMed] [Google Scholar]
- 5.Blackman, M. J., H. G. Heidrich, S. Donachie, J. S. McBride, and A. A. Holder. 1990. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J. Exp. Med. 172379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackman, M. J., and A. A. Holder. 1992. Secondary processing of the Plasmodium falciparum merozoite surface protein-1 (MSP1) by a calcium-dependent membrane-bound serine protease: shedding of MSP133 as a noncovalently associated complex with other fragments of the MSP1. Mol. Biochem. Parasitol. 50307-315. [DOI] [PubMed] [Google Scholar]
- 7.Blackman, M. J., I. T. Ling, S. C. Nicholls, and A. A. Holder. 1991. Proteolytic processing of the Plasmodium falciparum merozoite surface protein-1 produces a membrane-bound fragment containing two epidermal growth factor-like domains. Mol. Biochem. Parasitol. 4929-33. [DOI] [PubMed] [Google Scholar]
- 8.Blackman, M. J., T. J. Scott-Finnigan, S. Shai, and A. A. Holder. 1994. Antibodies inhibit the protease-mediated processing of a malaria merozoite surface protein. J. Exp. Med. 180389-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtidor, H., M. Urquiza, J. E. Suarez, L. E. Rodriguez, M. Ocampo, A. Puentes, J. E. Garcia, R. Vera, R. Lopez, L. E. Ramirez, M. Pinzon, and M. E. Patarroyo. 2001. Plasmodium falciparum acid basic repeat antigen (ABRA) peptides: erythrocyte binding and biological activity. Vaccine 194496-4504. [DOI] [PubMed] [Google Scholar]
- 10.Dolan, S. A., J. L. Proctor, D. W. Alling, Y. Okubo, T. E. Wellems, and L. H. Miller. 1994. Glycophorin B as an EBA-175 independent Plasmodium falciparum receptor of human erythrocytes. Mol. Biochem. Parasitol. 6455-63. [DOI] [PubMed] [Google Scholar]
- 11.Drew, D. R., R. A. O'Donnell, B. J. Smith, and B. S. Crabb. 2004. A common cross-species function for the double epidermal growth factor-like modules of the highly divergent plasmodium surface proteins MSP-1 and MSP-8. J. Biol. Chem. 27920147-20153. [DOI] [PubMed] [Google Scholar]
- 12.Drew, D. R., P. R. Sanders, and B. S. Crabb. 2005. Plasmodium falciparum merozoite surface protein 8 is a ring-stage membrane protein that localizes to the parasitophorous vacuole of infected erythrocytes. Infect. Immun. 733912-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duraisingh, M. T., T. Triglia, and A. F. Cowman. 2002. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int. J. Parasitol. 3281-89. [DOI] [PubMed] [Google Scholar]
- 14.Fidock, D. A., T. Nomura, and T. E. Wellems. 1998. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol. Pharmacol. 541140-1147. [DOI] [PubMed] [Google Scholar]
- 15.Florens, L., M. P. Washburn, J. D. Raine, R. M. Anthony, M. Grainger, J. D. Haynes, J. K. Moch, N. Muster, J. B. Sacci, D. L. Tabb, A. A. Witney, D. Wolters, Y. Wu, M. J. Gardner, A. A. Holder, R. E. Sinden, J. R. Yates, and D. J. Carucci. 2002. A proteomic view of the Plasmodium falciparum life cycle. Nature 419520-526. [DOI] [PubMed] [Google Scholar]
- 16.Galinski, M. R., A. R. Dluzewski, and J. W. Barnwell. 2005. A mechanistic approach to merozoite invasion of red blood cells::merozoite biogenesis, rupture, and invasion of erythrocytes, p. 113-168. In I. W. Sherman (ed.), Molecular approaches to malaria. ASM Press, Washington, DC.
- 17.Garcia, Y., A. Puentes, H. Curtidor, G. Cifuentes, C. Reyes, J. Barreto, A. Moreno, and M. E. Patarroyo. 2007. Identifying merozoite surface protein 4 and merozoite surface protein 7 Plasmodium falciparum protein family members specifically binding to human erythrocytes suggests a new malarial parasite-redundant survival mechanism. J. Med. Chem. 505665-5675. [DOI] [PubMed] [Google Scholar]
- 18.Gaur, D., D. C. Mayer, and L. H. Miller. 2004. Parasite ligand-host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int. J. Parasitol. 341413-1429. [DOI] [PubMed] [Google Scholar]
- 19.Gaur, D., J. R. Storry, M. E. Reid, J. W. Barnwell, and L. H. Miller. 2003. Plasmodium falciparum is able to invade erythrocytes through a trypsin-resistant pathway independent of glycophorin B. Infect. Immun. 716742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goel, V. K., X. Li, H. Chen, S. C. Liu, A. H. Chishti, and S. S. Oh. 2003. Band 3 is a host receptor binding merozoite surface protein 1 during the Plasmodium falciparum invasion of erythrocytes. Proc. Natl. Acad. Sci. USA 1005164-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayward, R. E., J. L. Derisi, S. Alfadhli, D. C. Kaslow, P. O. Brown, and P. K. Rathod. 2000. Shotgun DNA microarrays and stage-specific gene expression in Plasmodium falciparum malaria. Mol. Microbiol. 356-14. [DOI] [PubMed] [Google Scholar]
- 22.Holder, A. A., M. J. Blackman, P. A. Burghaus, J. A. Chappel, I. T. Ling, N. McCallum-Deighton, and S. Shai. 1992. A malaria merozoite surface protein (MSP1)—structure, processing and function. Mem. Inst. Oswaldo Cruz 87(Suppl. 3)37-42. [DOI] [PubMed] [Google Scholar]
- 23.Kauth, C. W., U. Woehlbier, M. Kern, Z. Mekonnen, R. Lutz, N. Mucke, J. Langowski, and H. Bujard. 2006. Interactions between merozoite surface proteins 1, 6, and 7 of the malaria parasite Plasmodium falciparum. J. Biol. Chem. 28131517-31527. [DOI] [PubMed] [Google Scholar]
- 24.Kushwaha, A., P. P. Rao, V. S. Duttu, P. Malhotra, and V. S. Chauhan. 2000. Expression and characterisation of Plasmodium falciparum acidic basic repeat antigen expressed in Escherichia coli. Mol. Biochem. Parasitol. 106213-224. [DOI] [PubMed] [Google Scholar]
- 25.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, P. De La Vega, A. A. Holder, S. Batalov, D. J. Carucci, and E. A. Winzeler. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 3011503-1508. [DOI] [PubMed] [Google Scholar]
- 26.Lopez, R., J. Valbuena, H. Curtidor, A. Puentes, L. E. Rodriguez, J. Garcia, J. Suarez, R. Vera, M. Ocampo, M. Trujillo, L. E. Ramirez, and M. E. Patarroyo. 2004. Plasmodium falciparum: red blood cell binding studies using peptides derived from rhoptry-associated protein 2 (RAP2). Biochimie 861-6. [DOI] [PubMed] [Google Scholar]
- 27.Mello, K., T. M. Daly, J. Morrisey, A. B. Vaidya, C. A. Long, and L. W. Bergman. 2002. A multigene family that interacts with the amino terminus of plasmodium MSP-1 identified using the yeast two-hybrid system. Eukaryot. Cell 1915-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills, K. E., J. A. Pearce, B. S. Crabb, and A. F. Cowman. 2002. Truncation of merozoite surface protein 3 disrupts its trafficking and that of acidic-basic repeat protein to the surface of Plasmodium falciparum merozoites. Mol. Microbiol. 431401-1411. [DOI] [PubMed] [Google Scholar]
- 29.Nikodem, D., and E. Davidson. 2000. Identification of a novel antigenic domain of Plasmodium falciparum merozoite surface protein-1 that specifically binds to human erythrocytes and inhibits parasite invasion, in vitro. Mol. Biochem. Parasitol. 10879-91. [DOI] [PubMed] [Google Scholar]
- 30.Okech, B., G. Mujuzi, A. Ogwal, H. Shirai, T. Horii, and T. G. Egwang. 2006. High titers of IgG antibodies against Plasmodium falciparum serine repeat antigen 5 (SERA5) are associated with protection against severe malaria in Ugandan children. Am. J. Trop. Med. Hyg. 74191-197. [PubMed] [Google Scholar]
- 31.Pachebat, J. A., M. Kadekoppala, M. Grainger, A. R. Dluzewski, R. S. Gunaratne, T. J. Scott-Finnigan, S. A. Ogun, I. T. Ling, L. H. Bannister, H. M. Taylor, G. H. Mitchell, and A. A. Holder. 2007. Extensive proteolytic processing of the malaria parasite merozoite surface protein 7 during biosynthesis and parasite release from erythrocytes. Mol. Biochem. Parasitol. 15159-69. [DOI] [PubMed] [Google Scholar]
- 32.Pachebat, J. A., I. T. Ling, M. Grainger, C. Trucco, S. Howell, D. Fernandez-Reyes, R. Gunaratne, and A. A. Holder. 2001. The 22 kDa component of the protein complex on the surface of Plasmodium falciparum merozoites is derived from a larger precursor, merozoite surface protein 7. Mol. Biochem. Parasitol. 11783-89. [DOI] [PubMed] [Google Scholar]
- 33.Pasvol, G., R. J. Wilson, M. E. Smalley, and J. Brown. 1978. Separation of viable schizont-infected red cells of Plasmodium falciparum from human blood. Ann. Trop. Med. Parasitol. 7287-88. [DOI] [PubMed] [Google Scholar]
- 34.Persson, K. E., C. T. Lee, K. Marsh, and J. G. Beeson. 2006. Development and optimization of high-throughput methods to measure Plasmodium falciparum-specific growth inhibitory antibodies. J. Clin. Microbiol. 441665-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rayner, J. C., M. R. Galinski, P. Ingravallo, and J. W. Barnwell. 2000. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc. Natl. Acad. Sci. USA 979648-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rayner, J. C., E. Vargas-Serrato, C. S. Huber, M. R. Galinski, and J. W. Barnwell. 2001. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J. Exp. Med. 1941571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez, L. E., H. Curtidor, M. Ocampo, J. Garcia, A. Puentes, J. Valbuena, R. Vera, R. Lopez, and M. E. Patarroyo. 2005. Identifying Plasmodium falciparum merozoite surface antigen 3 (MSP3) protein peptides that bind specifically to erythrocytes and inhibit merozoite invasion. Protein Sci. 141778-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders, P. R., G. T. Cantin, D. C. Greenbaum, P. R. Gilson, T. Nebl, R. L. Moritz, J. R. Yates III, A. N. Hodder, and B. S. Crabb. 2007. Identification of protein complexes in detergent-resistant membranes of Plasmodium falciparum schizonts. Mol. Biochem. Parasitol. 154148-157. [DOI] [PubMed] [Google Scholar]
- 39.Sanders, P. R., P. R. Gilson, G. T. Cantin, D. C. Greenbaum, T. Nebl, D. J. Carucci, M. J. McConville, L. Schofield, A. N. Hodder, J. R. Yates III, and B. S. Crabb. 2005. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J. Biol. Chem. 28040169-40176. [DOI] [PubMed] [Google Scholar]
- 40.Sanders, P. R., L. M. Kats, D. R. Drew, R. A. O'Donnell, M. O'Neill, A. G. Maier, R. L. Coppel, and B. S. Crabb. 2006. A set of glycosylphosphatidyl inositol-anchored membrane proteins of Plasmodium falciparum is refractory to genetic deletion. Infect. Immun. 744330-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stafford, W. H., M. J. Blackman, A. Harris, S. Shai, M. Grainger, and A. A. Holder. 1994. N-terminal amino acid sequence of the Plasmodium falciparum merozoite surface protein-1 polypeptides. Mol. Biochem. Parasitol. 66157-160. [DOI] [PubMed] [Google Scholar]
- 42.Stafford, W. H., B. Gunder, A. Harris, H. G. Heidrich, A. A. Holder, and M. J. Blackman. 1996. A 22 kDa protein associated with the Plasmodium falciparum merozoite surface protein-1 complex. Mol. Biochem. Parasitol. 80159-169. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, H. M., M. Grainger, and A. A. Holder. 2002. Variation in the expression of a Plasmodium falciparum protein family implicated in erythrocyte invasion. Infect. Immun. 705779-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tewari, R., S. A. Ogun, R. S. Gunaratne, A. Crisanti, and A. A. Holder. 2005. Disruption of Plasmodium berghei merozoite surface protein 7 gene modulates parasite growth in vivo. Blood 105394-396. [DOI] [PubMed] [Google Scholar]
- 45.Triglia, T., M. T. Duraisingh, R. T. Good, and A. F. Cowman. 2005. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol. Microbiol. 55162-174. [DOI] [PubMed] [Google Scholar]
- 46.Trucco, C., D. Fernandez-Reyes, S. Howell, W. H. Stafford, T. J. Scott-Finnigan, M. Grainger, S. A. Ogun, W. R. Taylor, and A. A. Holder. 2001. The merozoite surface protein 6 gene codes for a 36 kDa protein associated with the Plasmodium falciparum merozoite surface protein-1 complex. Mol. Biochem. Parasitol. 11291-101. [DOI] [PubMed] [Google Scholar]