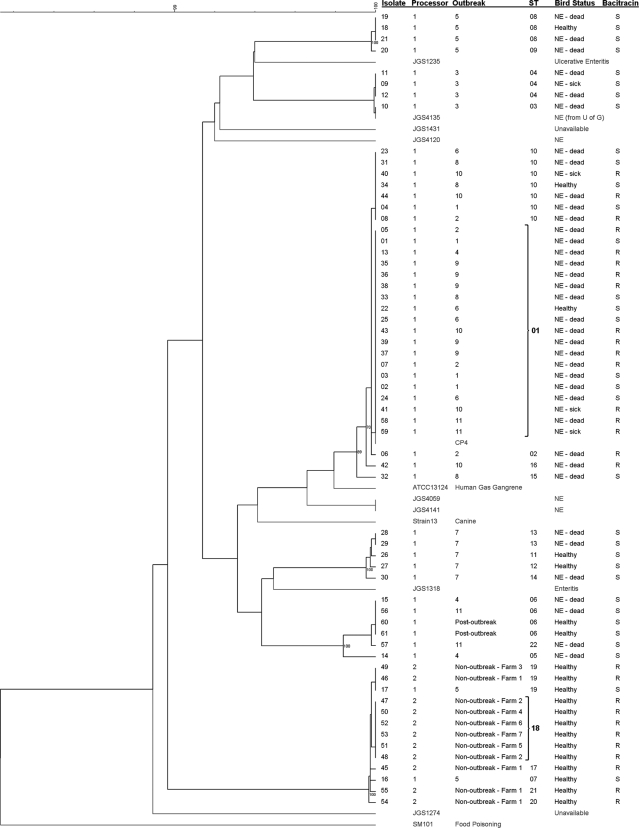
FIG. 1.
Dendrogram of concatenated MLST sequences of 61 avian isolates from Southern Ontario generated with pairwise similarities and UPGMA clustering. It includes eight previously published avian C. perfringens MLST sequences (16). Isolates from the study by Jost and collaborators are labeled JGS and shown in gray. CP4 is a virulent NE-associated isolate from another previous study in Ontario (30). The three C. perfringens strains with published genome sequences (strains 13, ATCC 13124, and SM101) were also included, and their MLST types were generated in silico (22, 27). Selected bootstrap values are indicated at nodes. Susceptibility to bacitracin was determined using a 16-μg/ml breakpoint. STs were determined through eBURSTv3 analysis (10), and ST01 and 18 are labeled in boldface type (founders).