Abstract
We have developed and evaluated a semiautomated assay for detection of nontuberculous mycobacteria (NTM) from clinical samples based on the Cobas Amplicor Mycobacterium tuberculosis test (Roche Diagnostics, Switzerland). A capture probe, specific for mycobacteria at the genus level, was linked to magnetic beads and used for the detection of amplification products obtained by the Cobas Amplicor M. tuberculosis assay. We demonstrate that the analytical sensitivity of the genus assay is similar to that of Cobas Amplicor M. tuberculosis detection. Four hundred sixteen clinical specimens were evaluated for the presence of NTM DNA. Sensitivities for smear-positive and smear-negative specimens were found to be 100% and 47.9%, respectively. Specificity was 97.7%, the positive predictive value 84.6%, and the negative predictive value 93.1%. The genus assay is easy to perform, produces reliable results, and was found to be a valuable diagnostic tool for rapid diagnosis of infections with NTM. The genus assay has the potential to detect NTM not routinely recovered by culture and to discover new mycobacterial species.
Nontuberculous mycobacteria (NTM) are frequently isolated from environmental sources, and about one-third of the species described have been associated with human disease (11, 16). Their pathogenic potential is variable, and infections preferentially occur in patients with underlying immunocompromising conditions, although immunocompetent individuals can also be affected. Clinical conditions caused by NTM include cutaneous ulcers, soft-tissue infections, lymphadenitis, joint infections, lung disease, and disseminated infections (11). Rapid laboratory diagnosis and differentiation from Mycobacterium tuberculosis are crucial, since treatment of NTM differs significantly from that of M. tuberculosis infections. Given the slow growth of most mycobacteria, fast and accurate molecular techniques are important for the diagnosis of suspected mycobacterial infections.
A variety of PCR-based test systems for direct detection of M. tuberculosis complex DNA from clinical specimens are commercially available and widely used, e.g., Cobas Amplicor M. tuberculosis (Roche Diagnostics, Switzerland) and AMTD2 (Gen-Probe) (15). In contrast, molecular genetic assays for direct detection of NTM are only infrequently implemented in routine diagnostics, although various in-house assays have been described (2, 4, 5, 8, 12, 21, 22). This is mainly due to the fact that existing assays are time-consuming and evaluation data are limited. Recently a commercial assay based on a manual line blot format was released for the direct detection of M. tuberculosis, M. avium, M. intracellulare, M. kansasii and M. malmoense from clinical samples (Hain Lifescience, Nehren, Germany) (7). However, numerous NTM well known to cause disease in humans are not included in this assay (e.g., M. marinum, M. abscessus, M. chelonae, M. xenopi, and M. ulcerans) (1, 6, 11, 14).
Here we present the development of an assay capable of detecting a large number of nontuberculous mycobacteria from clinical specimens using a semiautomated system well established for the detection of M. tuberculosis complex DNA.
MATERIALS AND METHODS
Decontamination of specimens, microscopy, and culture.
Specimens were decontaminated using the N-acetyl-l-cysteine-sodium hydroxide method for respiratory samples or the sodium hydroxide method for samples from sterile sites, following standard protocols (10). Microscopy was performed using auramine-rhodamine fluorochrome staining; positive microscopy results were confirmed using Ziehl-Neelsen staining (10). Standard media (Löwenstein-Jensen, 7H11 plates, and BBL MGIT [Becton, Dickinson and Company]) were used for cultural recovery, and cultures were incubated for 8 weeks at 37°C. For specimens from sterile sites, microscopy and inoculation of culture media were performed prior to decontamination. Mycobacteria grown in culture were identified by sequence analysis of the 16S rRNA gene as described previously (3, 12).
Clinical specimens.
Over a period of 3 months, the genus assay was performed with all samples for which molecular detection of M. tuberculosis was requested. In addition, we retrospectively included patient samples in the study which were positive for NTM by culture to allow calculation of sensitivity. Retrospectively analyzed samples were decontaminated and stored at 4°C prior to DNA extraction.
Although the Cobas Amplicor M. tuberculosis test is approved only for respiratory specimens, it is also widely used for nonrespiratory specimens (13, 17, 20, 24). Consequently, we included respiratory as well as nonrespiratory specimens in our study.
DNA extraction.
DNA was extracted from 0.5 ml of decontaminated specimen using the respiratory specimen preparation kit (Roche Diagnostics, Switzerland), following instructions for the Cobas Amplicor M. tuberculosis test (18).
Detection of M. tuberculosis complex DNA.
The Cobas Amplicor M. tuberculosis test was performed according to the manufacturer's instructions (Roche Diagnostics, Switzerland). Briefly, 50 μl master mix containing AmpliTaq DNA polymerase, biotinylated primers KY18 and K75, and an internal control plasmid were transferred into the detection ring, and 50 μl of DNA extract was added. The denaturation reagent, conjugate reagent, substrate, internal control detection reagent, and M. tuberculosis detection reagent were applied to the Cobas Amplicor analyzer as described in the operator's manual (18). Results were interpreted according to the manufacturer's instructions. A run was considered valid if the optical density at 660 nm (OD660) of the positive control was >2.0 and the OD660 of the negative control was <0.25. A sample was interpreted as positive if the OD660 was ≥0.35. A sample was interpreted as negative if the OD660 was <0.35 and the OD660 of the internal inhibition control was ≥0.35.
Labeling of magnetic beads with Mycobacterium genus-specific capture probe.
To adapt the Cobas Amplicor M. tuberculosis platform for the detection of nontuberculous mycobacteria, we constructed a Mycobacterium genus-specific capture probe. Magnetic beads harboring carboxylic acid groups on their surfaces were coupled to a Mycobacterium genus-specific oligonucleotide probe with an amino modification. Magnetic beads (Dynabeads M-270 carboxylic acid; Invitrogen, Germany) were washed four times with 25 mM 2-(N-morpholino)ethanesulfonic acid (MES) (Sigma-Aldrich Chemie GmbH, Germany) (pH 5) and incubated with oligonucleotide 259 (12) carrying an amino modification at the 5′end (5′ NH2-TTT CAC GAA CAA CGC GAC AA; tib molbiol, Germany) at a concentration of 2.1 nmol oligonucleotide per mg beads for at least 30 min at room temperature. Subsequently, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide HCl (EDC) (Sigma-Aldrich Chemie GmbH, Germany), resolved in cold 100 mM MES (pH 5), was added to the beads at a concentration of 10 mg per mg beads and incubated overnight at 4°C with shaking at 900 rpm. Labeled beads were washed four times using 50 mM Tris-HCl (pH 7.4) and stored in 50 mM Tris-HCl-0.1% Tween 20-0.02% sodium azide (pH 7.4) at 4°C until usage. All buffers were subjected to sterile filtration using a vacuum filtration system equipped with 0.22-μm membrane filters (Techno Plastic Product, Switzerland).
Detection of Mycobacterium genus DNA using Cobas Amplicor platform.
For detection of Mycobacterium genus DNA, we prepared a Mycobacterium genus-specific detection reagent. Beads labeled with the genus-specific probe (4.5 mg) were washed four times with 50 mM Tris (pH 7.4), resuspended in 1.3 ml of 25 mM MES-sodium azide (0.09%) (pH 9.18), and added to 6 ml of internal control buffer (Roche Diagnostics, Switzerland) in the original vial of the internal control buffer. The amount of beads used was adjusted by photometric measurement to an OD600 of 0.9 to 1.2.
The genus assay was performed on the Cobas Amplicor analyzer. For the detection of Mycobacterium genus DNA, the Mycobacterium genus detection reagent was applied to the analyzer instead of a detection reagent for M. tuberculosis DNA. A random barcode for an MTB detection vial was chosen in the Cobas analyzer software system, containing a “2” at the third position (MT2…), which indicated that the detection reagent was sufficient for 50 tests. This random barcode was then assigned to the Mycobacterium genus detection reagent. The rest of the procedure was identical to that of the Cobas Amplicor M. tuberculosis assay (18). To test a sample for the presence of both M. tuberculosis and Mycobacterium genus DNA, the Cobas Amplicor M. tuberculosis test was performed first. Subsequently, the ring containing the amplification products was transferred to the inner detection position and the same amplification products were detected again using the Mycobacterium genus detection reagent. The following algorithm was used for the interpretation of the genus assay. A run was considered valid if the OD660 of the positive control was >2.0 and the OD660 of the negative control was <0.5. A sample was considered positive if the OD660 was >0.5 and at least twofold higher than the background (negative control). A sample was considered negative if the OD660 was <0.5 and the OD660 of the internal control was >0.35. Samples with inhibition of amplification were excluded from the analysis.
Amplification, DNA purification, and sequencing of genus assay-positive samples.
Samples positive in the genus assay were amplified in a separate PCR to allow for sequence analysis without interference by the internal control plasmid. For additional PCRs, the same DNA extract was used. PCR amplification (95°C for 12 min and 40 cycles of 95°C for 1 min, 64°C for 1 min, 72°C for 2 min, and 72°C for 12 min) was performed in a total volume of 100 μl containing 5 U FastStart DNA polymerase, 1× PCR buffer with MgCl2 (Roche Diagnostics, Switzerland), 0.4 μM primer KY18 (5′-CAC ATG CAA GTC GAA CGG AAA GG; tib molbiol, Germany) (23), 0.4 μM primer KY75 (5′-GCC CGT ATC GCC CGC ACG CTC ACA) (23), 0.2 mM deoxynucleoside triphosphates (PCR nucleotide mix; Roche Diagnostics, Switzerland), and 20 μl of specimen DNA extract. Following purification of the PCR product (PCR purification kit, Qiagen, Germany) and cycle sequencing with primer Mbakt-14 (5′-GRG RTA CTC GAG TGG CGA AC [tib molbiol, Germany] and Avant 3100 [Applied Biosystems, Switzerland]), sequence analysis for species identification was performed as described previously (3). In the case of unsatisfactory sequence quality, the PCR product was reamplified (95°C for 12 min and 40 cycles of 95°C for 1 min, 58°C for 1 min, 72°C for 2 min, and 72°C for 12 min) using a seminested PCR approach with 2.5 U FastStart DNA polymerase including PCR buffer with MgCl2 (Roche Diagnostics, Switzerland), 0.4 μM primer KY18, 0.4 μM primer 259 (5′ TTT CAC GAA CAA CGC GAC AA; Tib Molbiol, Germany), 0.2 mM deoxynucleoside triphosphates (PCR nucleotide mix; Roche Diagnostics, Switzerland), and 2.5 μl PCR product in a total volume of 50 μl. If no PCR product could be obtained, amplification and reamplification were repeated as described above using alternative primer pairs 283 (5′-GAG TTT GAT CCT GGC TCA GGA, tib molbiol, Germany)/264 (5′-TGC ACA CAG GCC ACA AGG GA; tib molbiol, Germany) for amplification and primers 283/259 for reamplification. The resulting PCR product was subjected to purification, cycle sequencing, and sequence analysis as described above.
RESULTS
Establishment of direct detection assay for Mycobacterium genus DNA from clinical specimens.
We developed and validated an assay for direct detection of nontuberculous mycobacteria from clinical specimens using the Cobas Amplicor platform. Briefly, the Cobas Amplicor M. tuberculosis assay includes the following steps: (i) isolation of DNA from clinical samples, (ii) amplification of a 584-bp fragment of the 16S rRNA gene with biotinylated panmycobacterial primers (2, 23), (iii) capture of the PCR product with an M. tuberculosis-specific probe linked to magnetic beads, and (iv) photometric detection of the captured PCR product. The reaction mix contains an internal control plasmid to control for inhibition of the PCR. In order to use the Cobas Amplicor platform for the detection of nontuberculous mycobacteria, we generated a Mycobacterium genus-specific capture probe to be used as the specific detection reagent. Samples positive in the genus assay were amplified with panmycobacterial primers, and resulting amplification products were subjected to nucleic acid sequence determination to identify the Mycobacterium species.
Sensitivity of the genus assay compared to the Cobas Amplicor M. tuberculosis test.
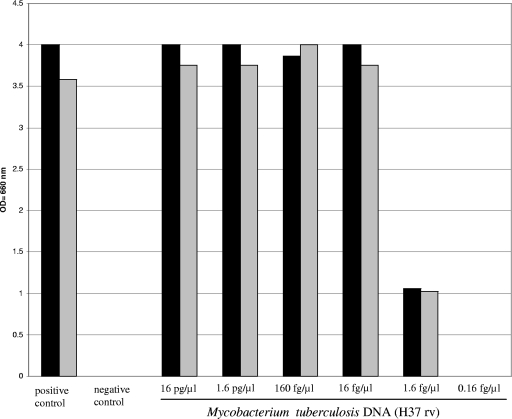
The analytical sensitivity of the genus assay was determined using a dilution series of M. tuberculosis DNA (H37RV) and compared to that of the Cobas Amplicor M. tuberculosis assay. The detection limit of both assays was 8 to 80 fg DNA per PCR, which is equivalent to approximately 2 to 20 genome copies (molecular weight of the M. tuberculosis genome, 4 fg) (9) (Fig. 1). Clinical evaluation showed that 15 of 431 specimens tested positive for the presence of M. tuberculosis DNA using the Cobas Amplicor M. tuberculosis assay. All 15 samples also gave a positive result using the genus assay (data not shown), further indicating that the genus assay is as sensitive as the commercial Cobas Amplicor M. tuberculosis assay.
FIG. 1.
Determination of analytical sensitivity of the genus assay in comparison to the Cobas Amplicor M. tuberculosis test using a dilution series of M. tuberculosis DNA. The detection limit of the genus assay (solid black) was similar to the detection limit of the Cobas Amplicor M. tuberculosis test (gray bars). The positive control and the negative control of the Cobas Amplicor M. tuberculosis test were included.
Evaluation of the genus assay with clinical specimens.
To evaluate the performance of the genus assay on clinical samples, the assay was incorporated into the routine diagnostic workflow. Samples screened for the presence of M. tuberculosis DNA were subsequently tested for the presence of NTM DNA using the genus assay. This strategy enabled us to screen a large number of clinical samples for the presence of NTM DNA using a semiautomated platform.
In total, 467 specimens were tested for the presence of M. tuberculosis and Mycobacterium genus DNA. This included 342 respiratory and 125 nonrespiratory specimens (46 puncture specimens, including aspirates from the pleural and abdominal cavities, 42 biopsies, 13 CSFs, 15 urines, 4 gastric aspirates, 3 stool samples, and 2 bone marrow specimens).
Inhibition of amplification as determined by negative internal control signals was detected in 36 of 467 (7.8%) specimens. In 15 (3.5%) of the remaining 431 samples, M. tuberculosis DNA was detected by both the genus assay and the Cobas Amplicor M. tuberculosis test (see above). These samples were excluded from the evaluation of the genus assay; thus, a total of 416 specimens were evaluated for the presence of NTM DNA.
In the genus assay, 52 of the 416 (12.5%) specimens tested positive, and the Mycobacterium species was further determined by sequence analysis. Samples were considered to be true positive in the genus assay if (i) the corresponding NTM was grown in culture, (ii) the corresponding NTM could be cultivated from other samples of the same patient, or (iii) the assay identified a noncultivable Mycobacterium species associated with a typical disease (e.g., M. leprae).
For 39 of 52 (75.0%) of the genus assay-positive samples species, identity was confirmed by culture (Table 1). Mycobacterial culture was negative for 13 samples positive in the genus assay. These discrepant samples were further analyzed to determine if NTM infection was present in the corresponding patients (Table 2). Three of the thirteen samples originated from patients with confirmed M. kansasii infections and were thus considered true positives. Two of the thirteen samples revealed the presence of M. leprae DNA; these samples were likewise considered true positives. For analysis of the remaining eight samples positive in the genus assay, see below.
TABLE 1.
Results of genus assay in comparison to those of culture and smear assays
| Smear test result | No. of samples with result
|
|||
|---|---|---|---|---|
| Genus assay positive
|
Genus assay negative
|
|||
| Culture positive | Culture negative | Culture positive | Culture negative | |
| Positive | 17 | 4 | 0 | 0 |
| Negative | 22 | 9 | 25 | 339 |
| Total | 39 | 13 | 25 | 339 |
TABLE 2.
Resolution of discrepant genus assay-positive, culture-negative results
| Mycobacterium genus assay result (OD660a) | Identification based on 16S rRNA gene sequence | Assay result
|
Specimen | Comment | |
|---|---|---|---|---|---|
| Smear | Culture | ||||
| Positive (2.344) | M. kansasii/gastri | Positive | No growth | Sputum | M. kansasii infectionb |
| Positive (1.177) | M. leprae | Positive | No growth | Biopsy | M. leprae infection |
| Positive (1.173) | M. leprae | Positive | No growth | Biopsy | M. leprae infection |
| Positive (3.935) | M. kansasii/gastri | Positive | No growth | Sputum | M. kansasii infectionb |
| Positive (1.335) | M. kansasii/gastri | Negative | No growth | Sputum | M. kansasii infectionb |
| Positive (0.604) | M. frederiksbergense | Negative | No growth | CSFd | Unknown significance |
| Positive (1.359) | M. smegmatis | Negative | No growth | Puncture fluid | Unknown significance |
| Positive (1.149) | M. fortuitum-complex | Negative | No growth | CSF | Unknown significance |
| Positive (1.059) | Corynebacterium sp. | Not done | No growth | Urine | False positive |
| Positive (0.928) | No sequencec | Negative | No growth | BALe | False positive |
| Positive (3.772) | No sequencec | Negative | No growth | BAL | False positive |
| Positive (>4.00) | No sequencec | Negative | No growth | Sputum | False positive |
| Positive (>4.00) | No sequencec | Negative | No growth | CSF | False positive |
OD660 of sample in genus assay.
Positive culture results for other specimens of patient.
No sequence could be obtained from the amplification product.
Cerebrospinal fluid.
Bronchoalveolar lavage.
For 64 specimens, culture revealed growth of nontuberculous mycobacteria (39 genus assay-positive samples and 25 genus assay-negative samples) (Table 1), resulting in a total number of 69 NTM-positive samples (Table 3). For smear-positive samples, the genus assay showed a sensitivity of 100% (21/21). For smear-negative samples, where the number of acid-fast bacilli can be as low as a single bacterium, the sensitivity was found to be 47.9% (23/48). Taking together smear-positive and -negative samples, the genus assay was able to detect 44/69 NTM-positive samples, resulting in an overall sensitivity of 63.8%. Species detected by the genus assay included M. kansasii, M. abscessus, M. chelonae, M. avium, M. intracellulare, M. xenopi, and M. leprae.
TABLE 3.
Performance of genus assay with samples positive or negative for NTM
| Smear test result | No. of samples with resulta
|
|||
|---|---|---|---|---|
| NTM positive
|
NTM negative
|
|||
| Genus assay positive | Genus assay negative | Genus assay positive | Genus assay negative | |
| Positive | 21 | 0 | 0 | 0 |
| Negative | 23 | 25 | 8 | 339 |
| Total | 44 | 25 | 8 | 339 |
A total of 69 samples were positive and 347 negative for NTM.
By culture, 347 samples were considered to be negative for NTM, including 8 genus assay-positive samples (Table 3). All of the eight genus assay-positive, culture-negative samples were smear negative. In one sample, sequence analysis revealed Corynebacterium sp., and in another three samples, fast-growing mycobacteria of uncertain clinical significance were identified. For the remaining four samples, sequence determination of the amplification product did not result in a clear sequence (Table 2).
Taken together, the specificity of the genus assay was 97.7%, the positive predictive value (PPV) 84.6%, and the negative predictive value (NPV) 93.1%. Sensitivity, specificity, PPV, and NPV determined for the Mycobacterium genus assay compare well with those published for the Cobas Amplicor M. tuberculosis test (15).
DISCUSSION
The possibility of genus-specific molecular assays in detection of mycobacterial infections has been recognized since the early 1990s (2). However, it has proven difficult, if not impossible, for routine clinical microbiological laboratories with little expertise in molecular diagnostics to implement homemade assays in their diagnostic workflow. To address this issue, we wished to develop a genus-specific molecular detection assay for mycobacteria on the basis of a widely available commercial instrumentation platform. Here we present the development and evaluation of a semiautomated assay for direct detection of nontuberculous mycobacteria from clinical samples. The Cobas Amplicor M. tuberculosis assay is well established in diagnostic laboratories. An assay based on this system has several advantages: (i) the basic test and platform are commercially available; (ii) a semiautomated detection system with little hands-on time allows high throughput, which is especially important in populations with a high proportion of negative samples; (iii) the PCR product obtained from the Cobas Amplicor M. tuberculosis test allows screening for both M. tuberculosis DNA and mycobacterial genus DNA; (iv) the positive control included in the Cobas Amplicor M. tuberculosis test can be used as a positive control for the genus assay (by using the same positive control, the performance of the genus assay can be directly compared to the Cobas Amplicor M. tuberculosis test for quality assurance); and (v) an internal control plasmid is included in the Cobas Amplicor M. tuberculosis test, which controls for inhibition of amplification.
To determine the analytical sensitivity of the genus assay, we tested a dilution series of M. tuberculosis DNA and demonstrated that there is no difference between the detection limit of the genus assay and that of the Cobas Amplicor M. tuberculosis assay (Fig. 1). The detection limit is in accordance with the published analytical sensitivity for purified DNA of five genome copies per PCR in 100% of the PCRs (18).
For evaluation of the genus assay in a routine diagnostic setting, 467 clinical samples were tested for the presence of mycobacterial DNA. These included 342 respiratory and 125 nonrespiratory specimens. The sensitivities of the genus assay were found to be 100%, 47.9%, and 63.8% for smear-positive samples, smear-negative samples, and overall sensitivity, respectively. These numbers are similar to the sensitivities published for the Cobas Amplicor M. tuberculosis test for extrapulmonary and respiratory specimens, ranging from 87.5 to 100% for smear-positive samples, from 17.2 to 70.8% for smear-negative samples, and from 27.3 to 85% for overall sensitivity, as reviewed by Piersimoni et al. (15). For respiratory specimens, the sensitivity for smear-positive and smear-negative samples and the overall sensitivity, as described in the manual of the manufacturer, are 96.4%, 72.8% and 86.6%, respectively (18). We conclude that the sensitivity of the genus assay is similar to that of the Cobas Amplicor M. tuberculosis test. The calculated overall sensitivity of the genus assay is relatively low in our study, since the number of smear-negative samples included in the study was more than twofold higher than that of smear-positive specimens tested.
A total of 52 samples were positive using the genus assay. Positive PCRs were confirmed by culture for 39 of the 52 samples. The remaining 13 samples were further analyzed to resolve the discrepancy (Table 2). For three (5.8%) genus assay-positive, culture-negative samples, sequence analysis of the PCR product revealed fast-growing mycobacteria of doubtful clinical significance. Given that fast-growing mycobacteria are found in various environmental sources, these results were considered most likely to represent contaminants. Since nontuberculous mycobacteria are frequently encountered in the environment, minimizing the risk of specimen contamination remains challenging. This not only involves assay setup but also includes all preanalytical steps (sample collection, transport, processing, decontamination, and DNA extraction). In one sample, Corynebacterium sp. was detected by nucleic acid analysis of the PCR product. The 16S rRNA sequences used as primer target sites in the Cobas Amplicor M. tuberculosis test are highly conserved within the genus Mycobacterium, including the majority of clinically relevant mycobacteria, with the exception of mycobacteria of the M. simiae group (19, 23). However, these 16S rRNA sequences are not exclusively specific for mycobacteria. Cross-reactivity with closely related bacteria, such as Corynebacterium species, Norcardia species, and Rhodococcus species, has been described (23). Cross-reactivity with these bacteria had only a minor impact on the performance of the genus assay, since all positive results were subjected to sequencing. Corynebacterium sp. was identified in 1/52 (1.9%) of genus assay-positive samples, indicating that cross-reactivity with these genera was a rather rare event in our evaluation. For four (7.7%) genus assay-positive specimens, sequence determination of the PCR product did not result in a defined sequence. Most likely this is a result of unspecific amplification in the genus assay.
In five (9.6%) genus assay-positive, culture-negative samples, we identified mycobacteria well established to cause disease in humans. Three samples revealed M. kansasii. All three samples were from patients with confirmed M. kansasii infections. Negative cultures are most likely due to intrasample variation and impaired viability of the organisms under antibiotic therapy. Two samples revealed the presence of M. leprae DNA; we consider these five samples to be correct positives in the genus assay.
Our results do point to the potential of the genus assay to detect mycobacteria which cannot be recovered by culture under normal laboratory conditions. This includes noncultivable mycobacteria (e.g., M. leprae), mycobacteria with special growth requirements (e.g., M. ulcerans), very slow-growing mycobacteria that exceed routine standard incubation times, and mycobacteria that fail to be cultivated, e.g., due to antibiotic therapy or harsh sample decontamination.
The specificity of the genus assay was 97.7%, the PPV 84.6%, and the NPV 93.1%. Again, these values are comparable to the described data for the Cobas Amplicor M. tuberculosis test, ranging from 91.3 to 100%, 73.3 to 100%, and 80.8 to 99.2%, respectively (15). The overall specificity, PPV, and NPV for respiratory specimens published in the manufacturer's manual are 99.7%, 96.3%, and 98.9%, respectively (18).
The eight false-positive samples in the genus assay were all smear negative and culture negative, and nucleic acid sequencing of the PCR product revealed sequences of fast-growing mycobacteria (n = 3) or Corynebacterium sp. (n = 1) or did not result in a defined sequence (n = 4) (Table 2). As a result of our evaluation, we report a positive genus assay result only if one of the following conditions is met: (i) the identified Mycobacterium species is clearly associated with disease; (ii) the sample is smear positive; or (iii) the same sequence has been obtained from other samples of the patient, which may include the laboratory request for additional analysis of samples in case only a single sample from a patient has been submitted to the laboratory. This strategy can increase the specificity and PPV to close to 100%, although it is difficult to determine true -positive and true-negative results when cultures remains negative and no additional reference method is available.
In conclusion, we have successfully developed a direct detection assay for NTM from clinical specimens based on the Cobas Amplicor M. tuberculosis platform. The genus assay and subsequent sequence analysis were able to identify the vast majority, 44/52 (84.6%), of NTM-positive samples correctly. The Mycobacterium species detected included M. kansasii, M. abscessus, M. chelonae, M. avium, M. intracellulare, M. xenopi, and M. leprae, which are all associated with human disease. The developed assay enables rapid and accurate diagnosis of infections with NTM, as well as recognition of unknown and noncultivable mycobacterial species.
Acknowledgments
This work was supported in part by the University of Zurich.
We thank Robert Schlaberg, Andreas Peter, and Nadine McCallum for critical reading of the manuscript.
We have no conflict of interest to disclose.
Footnotes
Published ahead of print on 22 October 2008.
REFERENCES
- 1.Andrejak, C., F. X. Lescure, Y. Douadi, G. Laurans, A. Smail, P. Duhaut, V. Jounieaux, and J. L. Schmit. 2007. Non-tuberculous mycobacteria pulmonary infection: management and follow-up of 31 infected patients. J. Infect. 5534-40. [DOI] [PubMed] [Google Scholar]
- 2.Böddinghaus, B., T. Rogall, T. Flohr, H. Blocker, and E. C. Böttger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 281751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosshard, P. P., R. Zbinden, S. Abels, B. Böddinghaus, M. Altwegg, and E. C. Böttger. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 441359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruijnesteijn Van Coppenraet, E. S., J. A. Lindeboom, J. M. Prins, M. F. Peeters, E. C. Claas, and E. J. Kuijper. 2004. Real-time PCR assay using fine-needle aspirates and tissue biopsy specimens for rapid diagnosis of mycobacterial lymphadenitis in children. J. Clin. Microbiol. 422644-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Portillo, P., M. C. Thomas, E. Martinez, C. Maranon, B. Valladares, M. E. Patarroyo, and M. Carlos Lopez. 1996. Multiprimer PCR system for differential identification of mycobacteria in clinical samples. J. Clin. Microbiol. 34324-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodiuk-Gad, R., P. Dyachenko, M. Ziv, A. Shani-Adir, Y. Oren, S. Mendelovici, J. Shafer, B. Chazan, R. Raz, Y. Keness, and D. Rozenman. 2007. Nontuberculous mycobacterial infections of the skin: a retrospective study of 25 cases. J. Am. Acad. Dermatol. 57413-420. [DOI] [PubMed] [Google Scholar]
- 7.Franco-Alvarez de Luna, F., P. Ruiz, J. Gutierrez, and M. Casal. 2006. Evaluation of the GenoType mycobacteria direct assay for detection of Mycobacterium tuberculosis complex and four atypical mycobacterial species in clinical samples. J. Clin. Microbiol. 443025-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Quintanilla, A., J. Gonzalez-Martin, G. Tudo, M. Espasa, and M. T. Jimenez de Anta. 2002. Simultaneous identification of Mycobacterium genus and Mycobacterium tuberculosis complex in clinical samples by 5′-exonuclease fluorogenic PCR. J. Clin. Microbiol. 404646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillemann, D., R. Warren, T. Kubica, S. Rusch-Gerdes, and S. Niemann. 2006. Rapid detection of Mycobacterium tuberculosis Beijing genotype strains by real-time PCR. J. Clin. Microbiol. 44302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isenberg, H. D. (ed.). 1992. Clinical microbiology procedures handbook, vol. 1. American Society for Microbiology, Washington, DC.
- 11.Katoch, V. M. 2004. Infections due to non-tuberculous mycobacteria (NTM). Indian J. Med. Res. 120290-304. [PubMed] [Google Scholar]
- 12.Kirschner, P., J. Rosenau, B. Springer, K. Teschner, K. Feldmann, and E. C. Böttger. 1996. Diagnosis of mycobacterial infections by nucleic acid amplification: 18-month prospective study. J. Clin. Microbiol. 34304-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitarai, S., H. Shishido, A. Kurashima, A. Tamura, and H. Nagai. 2000. Comparative study of Amplicor Mycobacterium PCR and conventional methods for the diagnosis of pleuritis caused by mycobacterial infection. Int. J. Tuberc. Lung Dis. 4871-876. [PubMed] [Google Scholar]
- 14.Petrini, B. 2006. Non-tuberculous mycobacterial infections. Scand. J. Infect. Dis. 38246-255. [DOI] [PubMed] [Google Scholar]
- 15.Piersimoni, C., and C. Scarparo. 2003. Relevance of commercial amplification methods for direct detection of Mycobacterium tuberculosis complex in clinical samples. J. Clin. Microbiol. 415355-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Primm, T. P., C. A. Lucero, and J. O. Falkinham III. 2004. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 1798-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reischl, U., N. Lehn, H. Wolf, and L. Naumann. 1998. Clinical evaluation of the automated COBAS AMPLICOR MTB assay for testing respiratory and nonrespiratory specimens. J. Clin. Microbiol. 362853-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roche Diagnostics. 2007. Cobas Amplicor Mycobacterium tuberculosis test: instruction manual. Roche Diagnostics, Mannheim, Germany.
- 19.Rogall, T., T. Flohr, and E. C. Böttger. 1990. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J. Gen. Microbiol. 1361915-1920. [DOI] [PubMed] [Google Scholar]
- 20.Shah, S., A. Miller, A. Mastellone, K. Kim, P. Colaninno, L. Hochstein, and R. D'Amato. 1998. Rapid diagnosis of tuberculosis in various biopsy and body fluid specimens by the AMPLICOR Mycobacterium tuberculosis polymerase chain reaction test. Chest 1131190-1194. [DOI] [PubMed] [Google Scholar]
- 21.Shrestha, N. K., M. J. Tuohy, G. S. Hall, U. Reischl, S. M. Gordon, and G. W. Procop. 2003. Detection and differentiation of Mycobacterium tuberculosis and nontuberculous mycobacterial isolates by real-time PCR. J. Clin. Microbiol. 415121-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stauffer, F., H. Haber, A. Rieger, R. Mutschlechner, P. Hasenberger, V. J. Tevere, and K. K. Young. 1998. Genus level identification of mycobacteria from clinical specimens by using an easy-to-handle Mycobacterium-specific PCR assay. J. Clin. Microbiol. 36614-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tevere, V. J., P. L. Hewitt, A. Dare, P. Hocknell, A. Keen, J. P. Spadoro, and K. K. Young. 1996. Detection of Mycobacterium tuberculosis by PCR amplification with pan-Mycobacterium primers and hybridization to an M. tuberculosis-specific probe. J. Clin. Microbiol. 34918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tortoli, E., M. Tronci, C. P. Tosi, C. Galli, F. Lavinia, S. Natili, and A. Goglio. 1999. Multicenter evaluation of two commercial amplification kits (Amplicor, Roche and LCx, Abbott) for direct detection of Mycobacterium tuberculosis in pulmonary and extrapulmonary specimens. Diagn. Microbiol. Infect. Dis. 33173-179. [DOI] [PubMed] [Google Scholar]