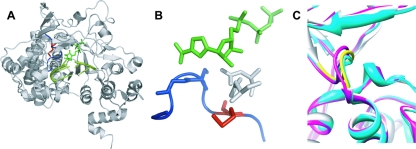
FIG. 1.
Structure of FMDV 3Dpol-RNA-RTP catalytic complex. (A) FMDV 3Dpol is shown in gray, the template-primer molecule is shown in light green, and the RTP molecule is represented by dark green sticks. Loop β9-α11 of FMDV 3Dpol is shown in blue, and the residue Met-296 is represented by red sticks. (B) A closer view of the β9-α11 loop (blue), with the 3Dpol residue Met-296 (red) and the RTP molecule (green). Blue sticks are residues Ser-298 and Gly-299, which establish interactions with the incoming nucleotide base (18). Asp-245 and Asn-307, which interact with the ribose of the incoming nucleotide, are represented by gray sticks. (C) Superimposition of structures of different FMDV 3Dpol complexes. Shown are the binary complex with template-primer RNA (Protein Database accession no. [pdb] 1WNE; gray); ternary complex with UTP/ATP (pdb 2EC0; magenta); and ternary complex with ribavirin (pdb 2E9R; cyan). The loop of residues 294 to 301 is shown in yellow. The distance between Cα atoms of Gly299 in the different complexes shown (taken as a measure of the movement of the loop of residues 297 to 301) is about 0.9 and 1.4 Å in the complexes with UTP/ATP and ribavirin, respectively.