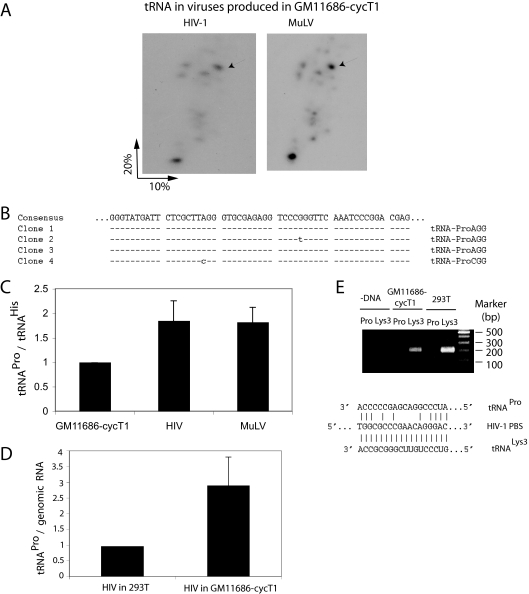
FIG. 3.
HIV-1 produced in murine cells packages tRNAPro. (A) 2D PAGE patterns of tRNAs in HIV-1 and MuLV, both produced in transiently transfected GM11686-cycT1 cells. (B) The numbered spots marked by arrows in Fig. 2A were extracted, amplified by RT-PCR using tRNAPro-specific primers, and sequenced. Clones 1 and 2 are MuLV, while clones 3 and 4 are HIV-1. (C) tRNAPro/tRNAHis ratios in GM11686-cycT1 cells and in the HIV-1 and MuLV produced in these cells. The tRNAPro/tRNAHis ratios were determined by hybridizing dot blots of cytoplasmic or viral RNA with probes specific for tRNAPro or tRNAHis. (D) tRNAPro/HIV-1 genomic RNA in HIV-1 produced in 293T cells or GM11686cyc-T1 cells, as determined by hybridizing dot blots of viral RNA with probes specific for tRNAPro and HIV-1 genomic RNA. (E) Identification of tRNA used to prime the synthesis of −SS DNA. SupT1 cells were infected with equal amounts of HIV-1 produced from either 293T cells or GM11686-cyc-T1 cells. DNA was extracted at different times postinfection, and equal amounts of DNA were amplified by PCR, using primer pairs complementary to −SS DNA (−DNA) and sequences near the 5′ terminus of either  (tRNALys3) or tRNAPro. (Top) PCR products were resolved by agarose gel electrophoresis. (Bottom) HIV-1 PBS sequence and sequences of the 3′ termini of tRNALys3 and tRNAPro.
(tRNALys3) or tRNAPro. (Top) PCR products were resolved by agarose gel electrophoresis. (Bottom) HIV-1 PBS sequence and sequences of the 3′ termini of tRNALys3 and tRNAPro.