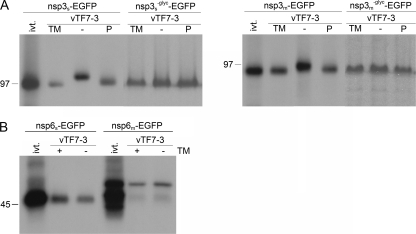
FIG. 2.
Processing of SARS-CoV and MHV nsp3 and nsp6. vTF7-3-infected OST7-1 cells were transfected with the indicated constructs. The cells were labeled with [35S]methionine from 5 to 6 h p.i., lysed, and processed for immunoprecipitation with antiserum directed to the EGFP tag, followed by SDS-PAGE. (A) Cells were transfected with SARS-CoV or MHV nsp3-EGFP (nsp3s-EGFP or nsp3m-EGFP, respectively)-encoding constructs without or with mutation (−glyc) of the N glycosylation sites in the presence (TM) or absence (− and P) of tunicamycin. The constructs with intact glycosylation sites were also transcribed and translated in vitro with the TNT coupled reticulocyte lysate system from Promega (ivt.). After immunoprecipitations, the samples were mock (TM, −) or PNGaseF (P) treated. (B) Cells were transfected with SARS-CoV or MHV nsp6-EGFP (nsp6s-EGFP and nsp6m-EGFP, respectively)-encoding constructs in the presence (+) or absence (−) of tunicamycin (TM). The same constructs were also transcribed and translated in vitro with the TNT coupled reticulocyte lysate system from Promega (ivt.). The positions and masses (in kilodaltons) of protein size markers are indicated at the left. Only the relevant portion of the gels is shown.